Check my LES run - 3D output#
The below notebook allows to compare a selected set of large-eddy simulation.
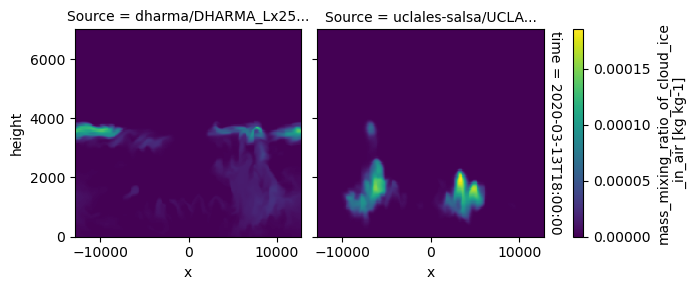
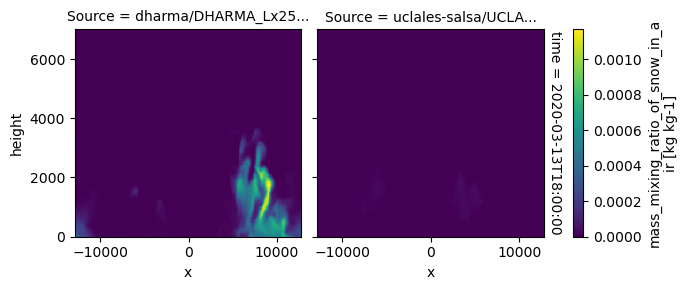
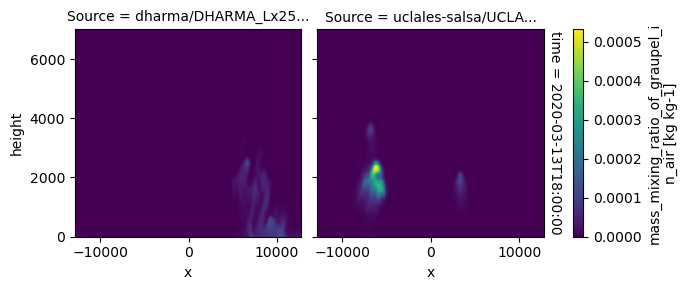
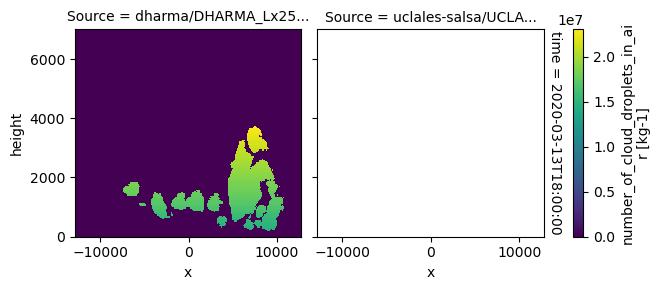
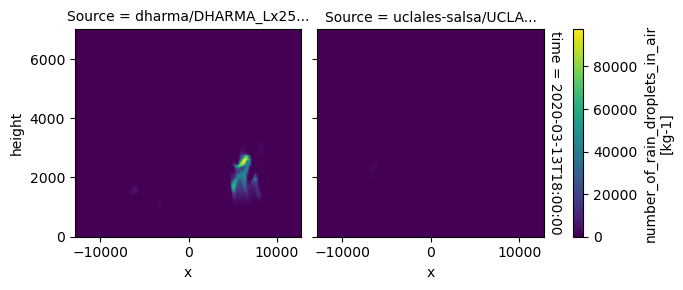
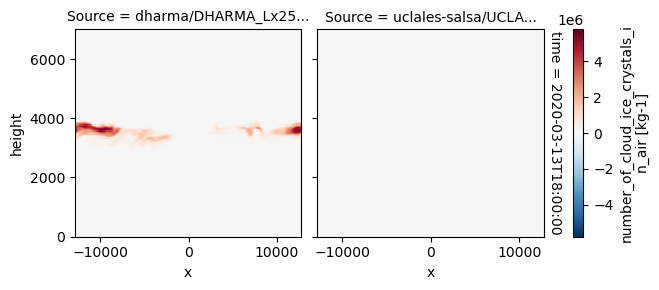
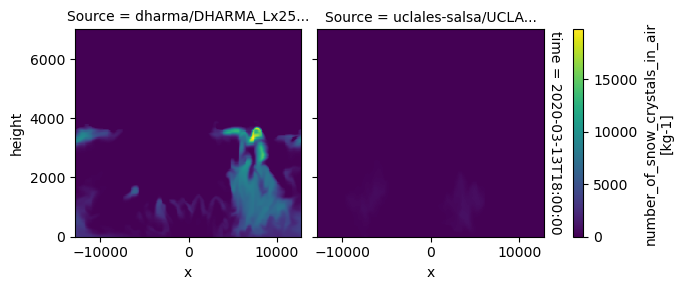
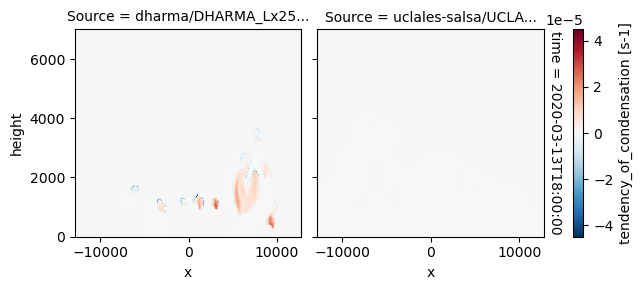
All requested 3D variables are here read and plotted. “load_sims_3d” reports variable that are missing. Maps will not show, if a variable is missing or if it contains missing values.
In case you find inconsistencies in the benchmark set or wish to expand the code, please contact Ann Fridlind (ann.fridlind@nasa.gov), Timothy Juliano (tjuliano@ucar.edu), and Florian Tornow (ft2544@columbia.edu).
%run functions_plotting.py
## select LES run(s) of interest
sim_keyword = 'DHARMA_Lx25_dx100_FixN_3D'
sim_keyword = 'FixN_3D'
## select specfic time
time_vec = [18.0]
y_sel = [100]
vars_mean_list = pd.read_excel('https://docs.google.com/spreadsheets/d/1Vl8jYGviet7EtXZuQiitrx4NSkV1x27aJAhxxjBb9zI/export?gid=1233994833&format=xlsx',
sheet_name='3D')
pd.set_option('display.max_rows', None)
vars_mean_list
## short names
var_vec_3d = np.asarray(vars_mean_list.variable_id[vars_mean_list.dimensions == 'time, height, x, y'])
## long names
var_vec_3d_long = np.asarray(vars_mean_list.standard_name[vars_mean_list.dimensions == 'time, height, x, y'])
## units
var_vec_3d_units = np.asarray(vars_mean_list.units[vars_mean_list.dimensions == 'time, height, x, y'])
%run functions_plotting.py
## load all simulations located in subfolders of the given directory
df_col_3d = load_sims_3d('/data/project/comble-mip/output_les/',var_vec_3d,t_shift=-2,times=time_vec,ysel=y_sel,keyword=sim_keyword)
Loading variables: f(time,height,x,y)
/data/project/comble-mip/output_les/dharma/DHARMA_Lx25_dx100_FixN_3D.nc
ERROR 1: PROJ: proj_create_from_database: Open of /opt/conda/share/proj failed
No field for na
No field for vmlc
No field for vmlr
No field for vmic
No field for vmis
No field for vmig
/data/project/comble-mip/output_les/uclales-salsa/UCLALES-SALSA_Lx25_dx100_FixN_3D.nc
...adjusting x and y values
Finding "zf" instead of "height"
NaN values in na
NaN values in nlc
NaN values in vmlc
NaN values in vmlr
NaN values in vmic
NaN values in vmis
NaN values in vmig
%run functions_plotting.py
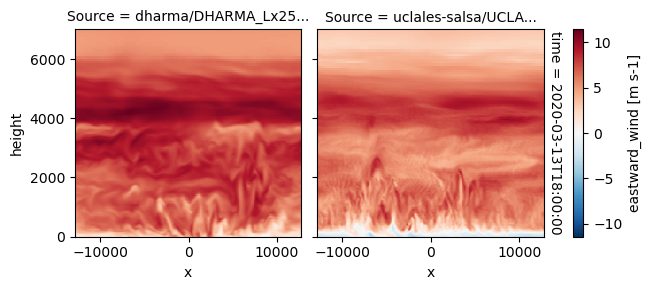
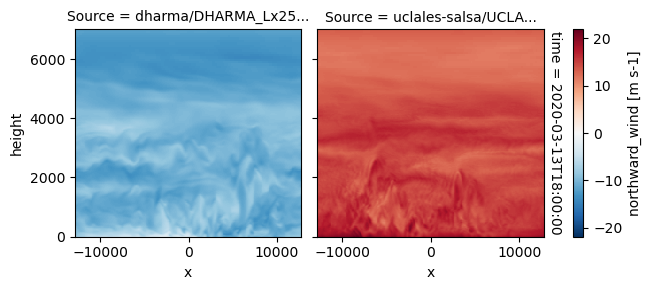
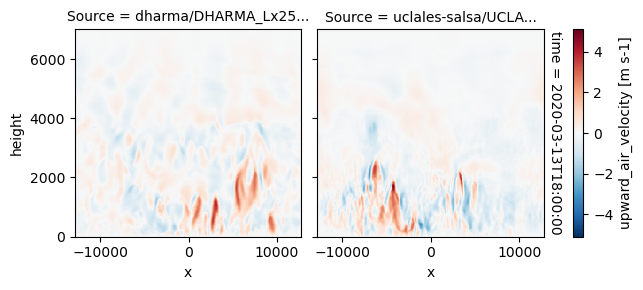
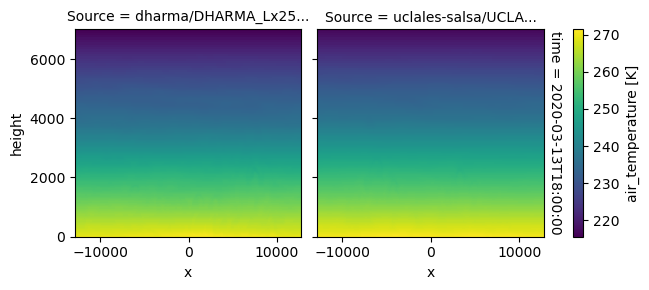
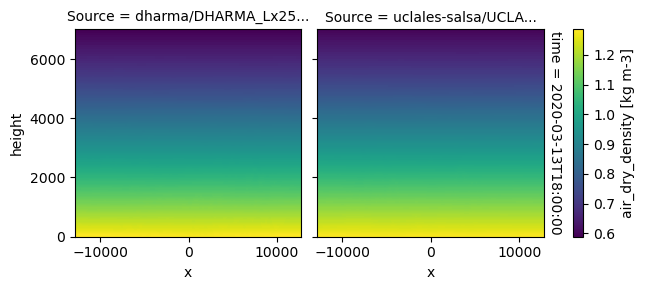
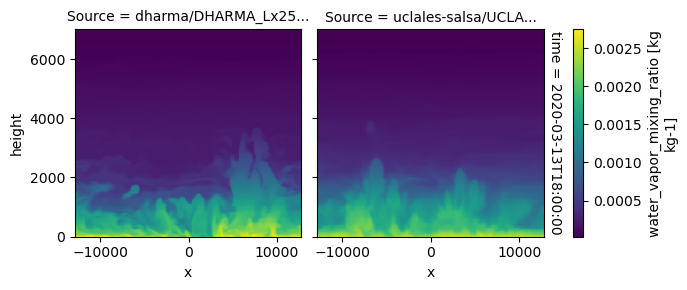
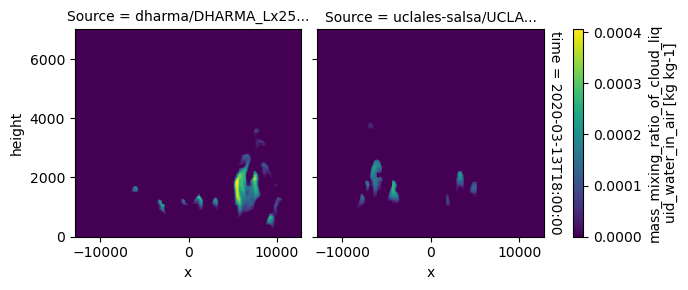
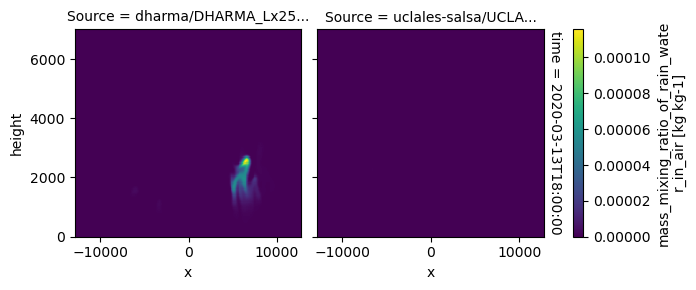
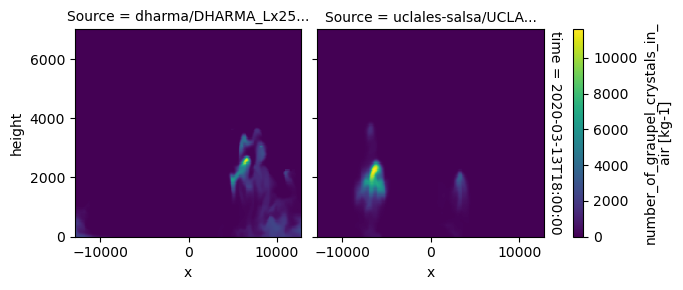
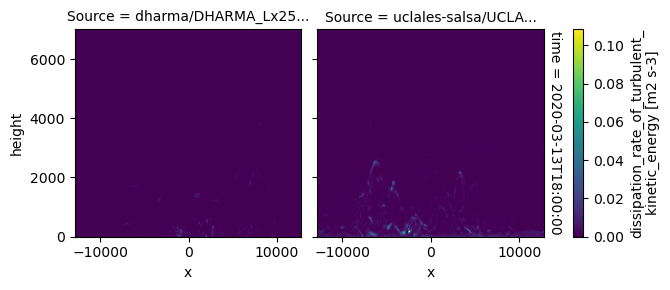
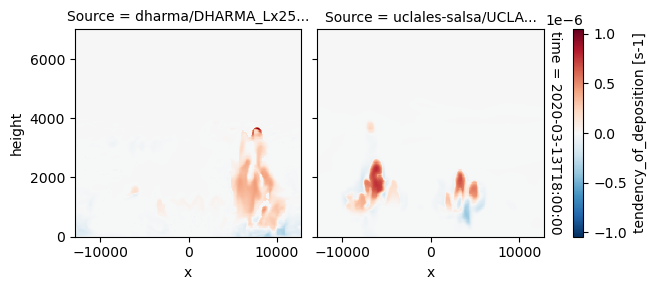
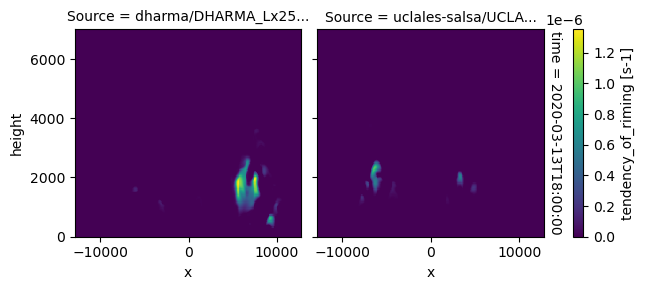
plot_2d_map(df_col_3d,var_vec_3d,times=time_vec)
/opt/conda/lib/python3.10/site-packages/xarray/plot/facetgrid.py:217: RuntimeWarning: More than 20 figures have been opened. Figures created through the pyplot interface (`matplotlib.pyplot.figure`) are retained until explicitly closed and may consume too much memory. (To control this warning, see the rcParam `figure.max_open_warning`). Consider using `matplotlib.pyplot.close()`.
fig, axs = plt.subplots(
<Figure size 1000x600 with 0 Axes>

