Example: convert DHARMA LES 2D output to DEPHY format#
Code to read DHARMA LES output files and write to DEPHY format (NetCDF)
Contributed by Florian Tornow Columbia University & NASA/GISS and Ann Fridlind from NASA/GISS
Import libraries#
import xarray as xr
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import csv
import os
import netCDF4
import datetime as dt
from netCDF4 import Dataset
import re
import sys
import pathlib
%run function_extract_2d.py
Specify directory locations#
If on the ARM JupyterHub, it is recommended to create and specify a local directory that is outside of the COMBLE-MIP repository to upload raw model output files in your model’s format.
Processed domain-mean outputs are invited for commit to the GitHub repository on a user-specified branch under /comble-mip/output_les/YOUR_MODEL_NAME/sandbox/YOUR_RUN_NAME. These can be committed and removed at any time.
It is requested to name your baseline small-domain run directory as ‘Lx25km_dx100m’ (in place of YOUR_RUN_NAME), so that it can readily be automatically compared with other runs using the same test specification.
# specify start time of simulation and simulation name
start_dtime = '2020-03-12 22:00:00.0'
# specify input data directory name, traceable to source machine, and
# specify output file name (see file naming convention in TOC)
# ProgNa with ice test
my_readdir = 'case0313_diag_ice25_nomiz_dx100_specZ0_hm_largedomain'
my_outfile = 'DHARMA_Lx25_dx100_FixN_2D_large.nc'
# FixN with no ice test
my_readdir = 'case0313_diag_ice0_nomiz_dx100_specZ0'
my_outfile = 'DHARMA_Lx25_dx100_FixN_noice_2D.nc'
my_readdir = 'case0313_prog_ice25_nomiz_dx100_specZ0_redo'
my_outfile = 'DHARMA_Lx25_dx100_ProgNa_2D.nc'
# specify local source directories (additional subdirectories if restart was required)
my_rundir = '/data/home/floriantornow/dharma/sandbox/' + my_readdir + '/'
my_outdirs = sorted([f for f in os.listdir(my_rundir) if not f.startswith('.')], key=str.lower)
print(my_outdirs)
# specify Github scratch directory where processed model output will be committed
my_gitdir = '../../output_les/dharma/sandbox/'
['0-20h']
Read DHARMA data#
Locate 2D files#
## locate all files that have the expected filename (*.tgz)
path = my_rundir + my_outdirs[0]
direc = pathlib.Path(path)
NCFILES = list(direc.rglob("*tgz"))
NCFILES_STR = [str(p) for p in pathlib.Path(path).rglob('*.tgz')]
print(NCFILES_STR)
input_filenames = []
count = 0
for fn in NCFILES:
if ('alt_0' in NCFILES_STR[count]):
input_filenames.append(fn)
count += 1
input_filenames = sorted(input_filenames)
['/data/home/floriantornow/dharma/sandbox/case0313_prog_ice25_nomiz_dx100_specZ0_redo/0-20h/dharma_PLT_007200.tgz', '/data/home/floriantornow/dharma/sandbox/case0313_prog_ice25_nomiz_dx100_specZ0_redo/0-20h/dharma_alt_3d_011524.tgz', '/data/home/floriantornow/dharma/sandbox/case0313_prog_ice25_nomiz_dx100_specZ0_redo/0-20h/dharma_alt_011524.tgz', '/data/home/floriantornow/dharma/sandbox/case0313_prog_ice25_nomiz_dx100_specZ0_redo/0-20h/dharma_PLT_011524.tgz', '/data/home/floriantornow/dharma/sandbox/case0313_prog_ice25_nomiz_dx100_specZ0_redo/0-20h/dharma_alt_3d_007200.tgz', '/data/home/floriantornow/dharma/sandbox/case0313_prog_ice25_nomiz_dx100_specZ0_redo/0-20h/dharma_alt_007200.tgz']
Read 2D files#
## obtain model horizontal resolution
input_filename = my_rundir + my_outdirs[0] + '/dharma.cdf'
dharma_params = xr.open_dataset(input_filename)
print(input_filename)
nx = dharma_params['geometry'].nx
Lx = dharma_params['geometry'].L_x
ERROR 1: PROJ: proj_create_from_database: Open of /opt/conda/share/proj failed
/data/home/floriantornow/dharma/sandbox/case0313_prog_ice25_nomiz_dx100_specZ0_redo/0-20h/dharma.cdf
xy_res = Lx/nx
xy_res
100.0
%run function_extract_2d.py
time_vec = []
counter = 0
for input_filename in input_filenames:
## obtain timestamp
time_vec.append(obtain_time(path,input_filename))
## ascertain horizontal resolution and prepare horizontal grid
xy_res = Lx/nx
x_vec = np.arange(-Lx/2, Lx/2, xy_res)
y_vec = np.arange(-Lx/2, Lx/2, xy_res)
## unpack tgz and extract relevant fields
opd_cloud = read_2d(FILE=input_filename,vbase = 'opd_drops',retain=True)
opd_ice = read_2d(FILE=input_filename,vbase = 'opd_ice',retain=True,there=True)
alb = (opd_cloud + opd_ice)/(13 + opd_cloud + opd_ice)
opd = opd_cloud + opd_ice
lwp = read_2d(FILE=input_filename,vbase = 'lwp',retain=True,there=True)/1000
iwp = read_2d(FILE=input_filename,vbase = 'iwp',retain=True,there=True)/1000
pflux = read_2d(FILE=input_filename,vbase = 'pflux',retain=True,there=True)
sfcTflx = read_2d(FILE=input_filename,vbase = 'sfc_T_flx',retain=True,there=True)
sfcQflx = read_2d(FILE=input_filename,vbase = 'sfc_qv_flx',retain=True,there=True)
u10m = read_2d(FILE=input_filename,vbase = 'u_layer1',retain=True,there=True) + dharma_params['translate'].u
v10m = read_2d(FILE=input_filename,vbase = 'v_layer1',retain=True,there=True) + dharma_params['translate'].v
LWup = read_2d(FILE=input_filename,vbase = 'Fup_toa_10um',there=True)
## accumulate
if counter == 0:
sfcTflx_col = sfcTflx
sfcQflx_col = sfcQflx
u10m_col = u10m
v10m_col = v10m
pflux_col = pflux
lwp_col = lwp
iwp_col = iwp
alb_col = alb
opd_col = opd
LWup_col = LWup
else:
sfcTflx_col = np.append(sfcTflx_col,sfcTflx,axis=0)
sfcQflx_col = np.append(sfcQflx_col,sfcQflx,axis=0)
u10m_col = np.append(u10m_col,u10m,axis=0)
v10m_col = np.append(v10m_col,v10m,axis=0)
pflux_col = np.append(pflux_col,pflux,axis=0)
lwp_col = np.append(lwp_col,lwp,axis=0)
iwp_col = np.append(iwp_col,iwp,axis=0)
alb_col = np.append(alb_col,alb,axis=0)
opd_col = np.append(opd_col,opd,axis=0)
LWup_col = np.append(LWup_col,LWup,axis=0)
counter += 1
| 7200 | 18000.0 | 1.0 | 0.3954E-09 | 0.6523E-11 | 0.4688 | 0.6000 |
| 11524 | 28800.0 | 1.1 | 0.1769E-09 | 0.4284E-12 | 0.0291 | 0.6000 |
## convert narrowband Flux (spectrally integrated) into Brightness Temperatureh = 6.62607015*(10**-34) ## J Hz-1
h = 6.62607015*(10**-34) ## J Hz-1
k = 1.380649*(10**-23) ## J⋅K−1
c = 2.99*(10**8) ## m s-1
lamb = 10.5*(10**-6)
BT_col = h*c/(k*lamb*np.log(1 + (2*h*(c**2)/((LWup_col/2.12/np.pi)*(10**6)*(lamb**5)))))
Prepare output file in DEPHY format#
Read requested variables list#
Variable description, naming, units, and dimensions.
# read list of requested variables
vars_mean_list = pd.read_excel('https://docs.google.com/spreadsheets/d/1Vl8jYGviet7EtXZuQiitrx4NSkV1x27aJAhxxjBb9zI/export?gid=1756539842&format=xlsx',
sheet_name='2D')
pd.set_option('display.max_rows', None)
vars_mean_list
| standard_name | variable_id | units | dimensions | comment (10-min instantaneous) | |
|---|---|---|---|---|---|
| 0 | time | time | s | – | dimension, seconds since 2020-03-12 18:00:00 |
| 1 | x | x | m | – | dimension, grid cell center |
| 2 | y | y | m | – | dimension, grid cell center |
| 3 | surface_upward_sensible_heat_flux | hfss | W m-2 | time, x, y | – |
| 4 | surface_upward_latent_heat_flux | hfls | W m-2 | time, x, y | – |
| 5 | surface_friction_velocity | ustar | m s-1 | time, x, y | – |
| 6 | surface_eastward_wind | us | m s-1 | time, x, y | at 10-m for comparison with SAR satellite meas... |
| 7 | surface_northward_wind | vs | m s-1 | time, x, y | at 10-m for comparison with SAR satellite meas... |
| 8 | precipitation_flux_at_surface | pr | kg m-2 s-1 | time, x, y | all hydrometeors |
| 9 | atmosphere_mass_content_of_liquid_water | lwp | kg m-2 | time, x, y | all liquid hydrometeors |
| 10 | atmosphere_mass_content_of_ice | iwp | kg m-2 | time, x, y | all ice hydrometeors |
| 11 | atmosphere_optical_thickness | opt | 1 | time, x, y | all hydrometeors for comparison with VIIRS; mi... |
| 12 | pseudo-albedo | alb | 1 | time, x, y | opt/(opt+13), where opt = all hydrometeor opti... |
| 13 | toa_outgoing_longwave_flux_atmospheric_window | olr11 | K | time, x, y | flux at top-of-atmosphere in window containing... |
shape(alb_col)
shape(lwp_col)
(2, 256, 256)
# create DEPHY output file
dephy_filename = './' + my_gitdir + my_outfile
if os.path.exists(dephy_filename):
os.remove(dephy_filename)
print('The file ' + dephy_filename + ' has been deleted successfully')
dephy_file = Dataset(dephy_filename,mode='w',format='NETCDF3_CLASSIC')
# create global attributes
dephy_file.title='DHARMA LES results for COMBLE-MIP case: fixed Nd and Ni'
dephy_file.reference='https://github.com/ARM-Development/comble-mip'
dephy_file.authors='Florian Tornow (florian.tornow@nasa.gov) and Ann Fridlind (ann.fridlind@nasa.gov)'
#dephy_file.source=input_filename
dephy_file.version=dt.datetime.now().strftime('%Y-%m-%d %H:%M:%S')
dephy_file.format_version='DEPHY-derivative'
dephy_file.script='convert_DHARMA_LES_output_to_dephy_format_2D.ipynb'
dephy_file.startDate=start_dtime
dephy_file.force_geo=1
dephy_file.surfaceType='ocean (after spin-up)'
dephy_file.surfaceForcing='ts (after spin-up)'
#dephy_file.lat=str(dharma_params['Coriolis'].lat) + ' deg N'
#dephy_file.dx=str(dharma_params['geometry'].L_x/dharma_params['geometry'].nx) + ' m'
#dephy_file.dy=str(dharma_params['geometry'].L_y/dharma_params['geometry'].ny) + ' m'
#dephy_file.dz='see zf variable'
#dephy_file.nx=dharma_params['geometry'].nx
#dephy_file.ny=dharma_params['geometry'].ny
#dephy_file.nz=dharma_params['geometry'].nz
# create dimensions
xvec = dephy_file.createDimension('x', nx)
xvec = dephy_file.createVariable('x', np.float64, ('x',))
xvec.units = 'm'
xvec.long_name = 'x'
xvec[:] = x_vec
yvec = dephy_file.createDimension('y', nx)
yvec = dephy_file.createVariable('y', np.float64, ('y',))
yvec.units = 'm'
yvec.long_name = 'y'
yvec[:] = y_vec
nt = len(input_filenames)
time = dephy_file.createDimension('time', nt)
time = dephy_file.createVariable('time', np.float64, ('time',))
time.units = 'seconds since ' + dephy_file.startDate
time.long_name = 'time'
time[:] = time_vec[:]
# create and fill variables
for index in vars_mean_list.index[3:]:
std_name = vars_mean_list.standard_name.iat[index]
var_name = vars_mean_list['variable_id'].iat[index]
#print(var_name)
if (var_name == 'hfss') | (var_name == 'hfls'):
continue
if var_name == 'hfss':
mod_field = sfcTflx_col
if var_name == 'hfls':
mod_field = sfcQflx_col
if var_name == 'us':
mod_field = u10m_col
if var_name == 'vs':
mod_field = v10m_col
if var_name == 'pr':
mod_field = pflux_col
if var_name == 'lwp':
mod_field = lwp_col
if var_name == 'iwp':
mod_field = iwp_col
if var_name == 'opt':
mod_field = opd_col
if var_name == 'alb':
mod_field = alb_col
if var_name == 'olr11':
mod_field = BT_col#LWup_col
if var_name == 'ustar':
continue
if vars_mean_list.dimensions.iat[index]=='time, x, y':
new_snd = dephy_file.createVariable(var_name, np.float64, ('time','x','y'))
new_snd.units = vars_mean_list.units.iat[index]
new_snd.long_name = std_name
#if vars_mean_list.model_name.iat[index]!='missing data': #and mod_name in dharma_snds:
new_snd[:] = mod_field
print(dephy_file)
dephy_file.close()
<class 'netCDF4._netCDF4.Dataset'>
root group (NETCDF3_CLASSIC data model, file format NETCDF3):
title: DHARMA LES results for COMBLE-MIP case: fixed Nd and Ni
reference: https://github.com/ARM-Development/comble-mip
authors: Florian Tornow (florian.tornow@nasa.gov) and Ann Fridlind (ann.fridlind@nasa.gov)
version: 2025-01-29 21:42:30
format_version: DEPHY-derivative
script: convert_DHARMA_LES_output_to_dephy_format_2D.ipynb
startDate: 2020-03-12 22:00:00.0
force_geo: 1
surfaceType: ocean (after spin-up)
surfaceForcing: ts (after spin-up)
dimensions(sizes): x(256), y(256), time(2)
variables(dimensions): float64 x(x), float64 y(y), float64 time(time), float64 us(time, x, y), float64 vs(time, x, y), float64 pr(time, x, y), float64 lwp(time, x, y), float64 iwp(time, x, y), float64 opt(time, x, y), float64 alb(time, x, y), float64 olr11(time, x, y)
groups:
dephy_check = xr.open_dataset(dephy_filename)
dephy_check
<xarray.Dataset> Size: 8MB
Dimensions: (x: 256, y: 256, time: 2)
Coordinates:
* x (x) float64 2kB -1.28e+04 -1.27e+04 -1.26e+04 ... 1.26e+04 1.27e+04
* y (y) float64 2kB -1.28e+04 -1.27e+04 -1.26e+04 ... 1.26e+04 1.27e+04
* time (time) datetime64[ns] 16B 2020-03-13T03:00:00 2020-03-13T06:00:00
Data variables:
us (time, x, y) float64 1MB ...
vs (time, x, y) float64 1MB ...
pr (time, x, y) float64 1MB ...
lwp (time, x, y) float64 1MB ...
iwp (time, x, y) float64 1MB ...
opt (time, x, y) float64 1MB ...
alb (time, x, y) float64 1MB ...
olr11 (time, x, y) float64 1MB ...
Attributes:
title: DHARMA LES results for COMBLE-MIP case: fixed Nd and Ni
reference: https://github.com/ARM-Development/comble-mip
authors: Florian Tornow (florian.tornow@nasa.gov) and Ann Fridlin...
version: 2025-01-29 21:42:30
format_version: DEPHY-derivative
script: convert_DHARMA_LES_output_to_dephy_format_2D.ipynb
startDate: 2020-03-12 22:00:00.0
force_geo: 1
surfaceType: ocean (after spin-up)
surfaceForcing: ts (after spin-up)dephy_filename
'./../../output_les/dharma/sandbox/DHARMA_Lx25_dx100_ProgNa_2D.nc'
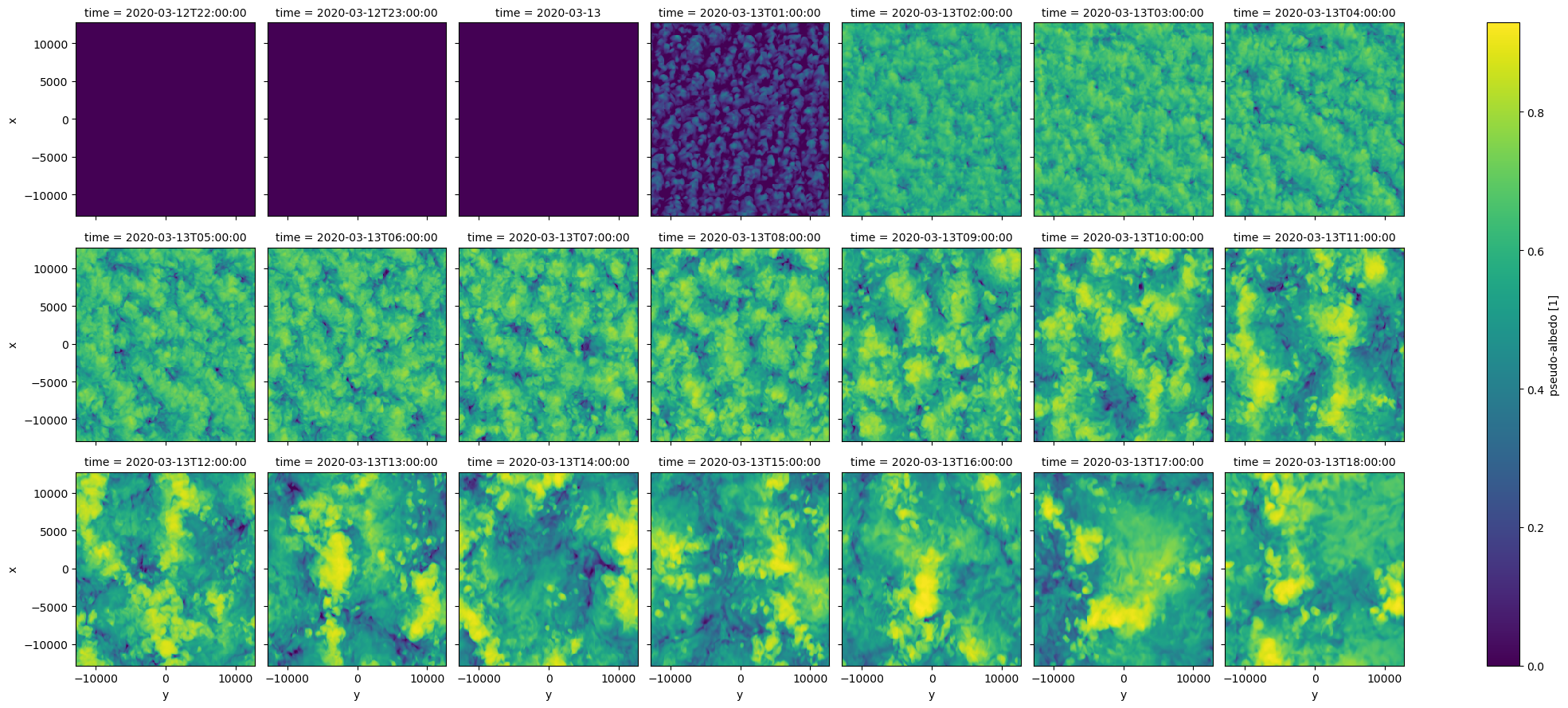
#dephy_check['olr11'].plot(row='time')
dephy_check['alb'].plot(row='time',col_wrap=7)
<xarray.plot.facetgrid.FacetGrid at 0x7f65f39ec220>