
Introduction to Xarray
Overview
This notebook will introduce the basics of gridded, labeled data with Xarray. Since Xarray introduces additional abstractions on top of plain arrays of data, our goal is to show why these abstractions are useful and how they frequently lead to simpler, more robust code.
We’ll cover these topics:
Create a
DataArray, one of the core object types in XarrayUnderstand how to use named coordinates and metadata in a
DataArrayCombine individual
DataArraysinto aDataset, the other core object type in XarraySubset, slice, and interpolate the data using named coordinates
Open netCDF data using XArray
Basic subsetting and aggregation of a
DatasetBrief introduction to plotting with Xarray
Prerequisites
Concepts |
Importance |
Notes |
|---|---|---|
Necessary |
||
Helpful |
Familiarity with indexing and slicing arrays |
|
Helpful |
Familiar with array arithmetic and broadcasting |
|
Helpful |
Familiarity with labeled data |
|
Helpful |
Familiarity with time formats and the |
|
Helpful |
Familiarity with metadata structure |
Time to learn: 30 minutes
Imports
Simmilar to numpy, np; pandas, pd; you may often encounter xarray imported within a shortened namespace as xr.
from datetime import timedelta
import cmweather
import numpy as np
import pandas as pd
import xarray as xr
import glob
from bokeh.models.formatters import DatetimeTickFormatter
import hvplot.xarray
import holoviews as hv
hv.extension("bokeh")
Introducing the DataArray and Dataset
Xarray expands on the capabilities on NumPy arrays, providing a lot of streamlined data manipulation. It is similar in that respect to Pandas, but whereas Pandas excels at working with tabular data, Xarray is focused on N-dimensional arrays of data (i.e. grids). Its interface is based largely on the netCDF data model (variables, attributes, and dimensions), but it goes beyond the traditional netCDF interfaces to provide functionality similar to netCDF-java’s Common Data Model (CDM).
Creation of a DataArray object
The DataArray is one of the basic building blocks of Xarray (see docs here). It provides a numpy.ndarray-like object that expands to provide two critical pieces of functionality:
Coordinate names and values are stored with the data, making slicing and indexing much more powerful
It has a built-in container for attributes
Here we’ll initialize a DataArray object by wrapping a plain NumPy array, and explore a few of its properties.
Generate a random numpy array
For our first example, we’ll just create a random array of “temperature” data in units of Kelvin:
data = 283 + 5 * np.random.randn(5, 3, 4)
data
array([[[286.27956776, 277.95199244, 282.03019204, 289.72997186],
[291.40837238, 280.71852405, 288.30962373, 278.38771859],
[283.54050134, 281.50088874, 283.67147977, 282.69508194]],
[[275.53284611, 285.37426651, 275.88078048, 285.61579451],
[290.57368097, 280.81104924, 275.86434679, 280.78811485],
[278.07454137, 290.51130363, 291.4245341 , 284.55193302]],
[[276.37392534, 285.66779544, 275.7531373 , 285.74384742],
[285.6877712 , 283.67698247, 281.84754872, 276.55362898],
[285.59721942, 276.05622785, 286.45987133, 289.96323162]],
[[270.92446753, 282.98749258, 284.09955379, 284.50343734],
[277.34994456, 278.27143906, 277.69850219, 284.11487834],
[275.94418059, 283.27421098, 269.83904741, 276.73236976]],
[[282.15740785, 286.21591172, 279.85464686, 285.96992707],
[290.88477194, 281.37684085, 281.49092035, 282.16621324],
[283.85810564, 285.68428321, 277.78787953, 285.17050016]]])
Wrap the array: first attempt
Now we create a basic DataArray just by passing our plain data as input:
temp = xr.DataArray(data)
temp
<xarray.DataArray (dim_0: 5, dim_1: 3, dim_2: 4)>
array([[[286.27956776, 277.95199244, 282.03019204, 289.72997186],
[291.40837238, 280.71852405, 288.30962373, 278.38771859],
[283.54050134, 281.50088874, 283.67147977, 282.69508194]],
[[275.53284611, 285.37426651, 275.88078048, 285.61579451],
[290.57368097, 280.81104924, 275.86434679, 280.78811485],
[278.07454137, 290.51130363, 291.4245341 , 284.55193302]],
[[276.37392534, 285.66779544, 275.7531373 , 285.74384742],
[285.6877712 , 283.67698247, 281.84754872, 276.55362898],
[285.59721942, 276.05622785, 286.45987133, 289.96323162]],
[[270.92446753, 282.98749258, 284.09955379, 284.50343734],
[277.34994456, 278.27143906, 277.69850219, 284.11487834],
[275.94418059, 283.27421098, 269.83904741, 276.73236976]],
[[282.15740785, 286.21591172, 279.85464686, 285.96992707],
[290.88477194, 281.37684085, 281.49092035, 282.16621324],
[283.85810564, 285.68428321, 277.78787953, 285.17050016]]])
Dimensions without coordinates: dim_0, dim_1, dim_2Note two things:
Xarray generates some basic dimension names for us (
dim_0,dim_1,dim_2). We’ll improve this with better names in the next example.Wrapping the numpy array in a
DataArraygives us a rich display in the notebook! (Try clicking the array symbol to expand or collapse the view)
Assign dimension names
Much of the power of Xarray comes from making use of named dimensions. So let’s add some more useful names! We can do that by passing an ordered list of names using the keyword argument dims:
temp = xr.DataArray(data, dims=['time', 'lat', 'lon'])
temp
<xarray.DataArray (time: 5, lat: 3, lon: 4)>
array([[[286.27956776, 277.95199244, 282.03019204, 289.72997186],
[291.40837238, 280.71852405, 288.30962373, 278.38771859],
[283.54050134, 281.50088874, 283.67147977, 282.69508194]],
[[275.53284611, 285.37426651, 275.88078048, 285.61579451],
[290.57368097, 280.81104924, 275.86434679, 280.78811485],
[278.07454137, 290.51130363, 291.4245341 , 284.55193302]],
[[276.37392534, 285.66779544, 275.7531373 , 285.74384742],
[285.6877712 , 283.67698247, 281.84754872, 276.55362898],
[285.59721942, 276.05622785, 286.45987133, 289.96323162]],
[[270.92446753, 282.98749258, 284.09955379, 284.50343734],
[277.34994456, 278.27143906, 277.69850219, 284.11487834],
[275.94418059, 283.27421098, 269.83904741, 276.73236976]],
[[282.15740785, 286.21591172, 279.85464686, 285.96992707],
[290.88477194, 281.37684085, 281.49092035, 282.16621324],
[283.85810564, 285.68428321, 277.78787953, 285.17050016]]])
Dimensions without coordinates: time, lat, lonThis is already improved upon from a NumPy array, because we have names for each of the dimensions (or axes in NumPy parlance). Even better, we can take arrays representing the values for the coordinates for each of these dimensions and associate them with the data when we create the DataArray. We’ll see this in the next example.
Create a DataArray with named Coordinates
Make time and space coordinates
Here we will use Pandas to create an array of datetime data, which we will then use to create a DataArray with a named coordinate time.
times = pd.date_range('2018-01-01', periods=5)
times
DatetimeIndex(['2018-01-01', '2018-01-02', '2018-01-03', '2018-01-04',
'2018-01-05'],
dtype='datetime64[ns]', freq='D')
We’ll also create arrays to represent sample longitude and latitude:
lons = np.linspace(-120, -60, 4)
lats = np.linspace(25, 55, 3)
Initialize the DataArray with complete coordinate info
When we create the DataArray instance, we pass in the arrays we just created:
temp = xr.DataArray(data, coords=[times, lats, lons], dims=['time', 'lat', 'lon'])
temp
<xarray.DataArray (time: 5, lat: 3, lon: 4)>
array([[[286.27956776, 277.95199244, 282.03019204, 289.72997186],
[291.40837238, 280.71852405, 288.30962373, 278.38771859],
[283.54050134, 281.50088874, 283.67147977, 282.69508194]],
[[275.53284611, 285.37426651, 275.88078048, 285.61579451],
[290.57368097, 280.81104924, 275.86434679, 280.78811485],
[278.07454137, 290.51130363, 291.4245341 , 284.55193302]],
[[276.37392534, 285.66779544, 275.7531373 , 285.74384742],
[285.6877712 , 283.67698247, 281.84754872, 276.55362898],
[285.59721942, 276.05622785, 286.45987133, 289.96323162]],
[[270.92446753, 282.98749258, 284.09955379, 284.50343734],
[277.34994456, 278.27143906, 277.69850219, 284.11487834],
[275.94418059, 283.27421098, 269.83904741, 276.73236976]],
[[282.15740785, 286.21591172, 279.85464686, 285.96992707],
[290.88477194, 281.37684085, 281.49092035, 282.16621324],
[283.85810564, 285.68428321, 277.78787953, 285.17050016]]])
Coordinates:
* time (time) datetime64[ns] 2018-01-01 2018-01-02 ... 2018-01-05
* lat (lat) float64 25.0 40.0 55.0
* lon (lon) float64 -120.0 -100.0 -80.0 -60.0Set useful attributes
…and while we’re at it, we can also set some attribute metadata:
temp.attrs['units'] = 'kelvin'
temp.attrs['standard_name'] = 'air_temperature'
temp
<xarray.DataArray (time: 5, lat: 3, lon: 4)>
array([[[286.27956776, 277.95199244, 282.03019204, 289.72997186],
[291.40837238, 280.71852405, 288.30962373, 278.38771859],
[283.54050134, 281.50088874, 283.67147977, 282.69508194]],
[[275.53284611, 285.37426651, 275.88078048, 285.61579451],
[290.57368097, 280.81104924, 275.86434679, 280.78811485],
[278.07454137, 290.51130363, 291.4245341 , 284.55193302]],
[[276.37392534, 285.66779544, 275.7531373 , 285.74384742],
[285.6877712 , 283.67698247, 281.84754872, 276.55362898],
[285.59721942, 276.05622785, 286.45987133, 289.96323162]],
[[270.92446753, 282.98749258, 284.09955379, 284.50343734],
[277.34994456, 278.27143906, 277.69850219, 284.11487834],
[275.94418059, 283.27421098, 269.83904741, 276.73236976]],
[[282.15740785, 286.21591172, 279.85464686, 285.96992707],
[290.88477194, 281.37684085, 281.49092035, 282.16621324],
[283.85810564, 285.68428321, 277.78787953, 285.17050016]]])
Coordinates:
* time (time) datetime64[ns] 2018-01-01 2018-01-02 ... 2018-01-05
* lat (lat) float64 25.0 40.0 55.0
* lon (lon) float64 -120.0 -100.0 -80.0 -60.0
Attributes:
units: kelvin
standard_name: air_temperatureAttributes are not preserved by default!
Notice what happens if we perform a mathematical operaton with the DataArray: the coordinate values persist, but the attributes are lost. This is done because it is very challenging to know if the attribute metadata is still correct or appropriate after arbitrary arithmetic operations.
To illustrate this, we’ll do a simple unit conversion from Kelvin to Celsius:
temp_in_celsius = temp - 273.15
temp_in_celsius
<xarray.DataArray (time: 5, lat: 3, lon: 4)>
array([[[13.12956776, 4.80199244, 8.88019204, 16.57997186],
[18.25837238, 7.56852405, 15.15962373, 5.23771859],
[10.39050134, 8.35088874, 10.52147977, 9.54508194]],
[[ 2.38284611, 12.22426651, 2.73078048, 12.46579451],
[17.42368097, 7.66104924, 2.71434679, 7.63811485],
[ 4.92454137, 17.36130363, 18.2745341 , 11.40193302]],
[[ 3.22392534, 12.51779544, 2.6031373 , 12.59384742],
[12.5377712 , 10.52698247, 8.69754872, 3.40362898],
[12.44721942, 2.90622785, 13.30987133, 16.81323162]],
[[-2.22553247, 9.83749258, 10.94955379, 11.35343734],
[ 4.19994456, 5.12143906, 4.54850219, 10.96487834],
[ 2.79418059, 10.12421098, -3.31095259, 3.58236976]],
[[ 9.00740785, 13.06591172, 6.70464686, 12.81992707],
[17.73477194, 8.22684085, 8.34092035, 9.01621324],
[10.70810564, 12.53428321, 4.63787953, 12.02050016]]])
Coordinates:
* time (time) datetime64[ns] 2018-01-01 2018-01-02 ... 2018-01-05
* lat (lat) float64 25.0 40.0 55.0
* lon (lon) float64 -120.0 -100.0 -80.0 -60.0For an in-depth discussion of how Xarray handles metadata, start in the Xarray docs here.
Subsetting and selection by coordinate values
Much of the power of labeled coordinates comes from the ability to select data based on coordinate names and values, rather than array indices. We’ll explore this briefly here.
NumPy-like selection
Suppose we want to extract all the spatial data for one single date: January 2, 2018. It’s possible to achieve that with NumPy-like index selection:
indexed_selection = temp[1, :, :] # Index 1 along axis 0 is the time slice we want...
indexed_selection
<xarray.DataArray (lat: 3, lon: 4)>
array([[275.53284611, 285.37426651, 275.88078048, 285.61579451],
[290.57368097, 280.81104924, 275.86434679, 280.78811485],
[278.07454137, 290.51130363, 291.4245341 , 284.55193302]])
Coordinates:
time datetime64[ns] 2018-01-02
* lat (lat) float64 25.0 40.0 55.0
* lon (lon) float64 -120.0 -100.0 -80.0 -60.0
Attributes:
units: kelvin
standard_name: air_temperatureHOWEVER, notice that this requires us (the user / programmer) to have detailed knowledge of the order of the axes and the meaning of the indices along those axes!
Named coordinates free us from this burden…
Selecting with .sel()
We can instead select data based on coordinate values using the .sel() method, which takes one or more named coordinate(s) as keyword argument:
named_selection = temp.sel(time='2018-01-02')
named_selection
<xarray.DataArray (lat: 3, lon: 4)>
array([[275.53284611, 285.37426651, 275.88078048, 285.61579451],
[290.57368097, 280.81104924, 275.86434679, 280.78811485],
[278.07454137, 290.51130363, 291.4245341 , 284.55193302]])
Coordinates:
time datetime64[ns] 2018-01-02
* lat (lat) float64 25.0 40.0 55.0
* lon (lon) float64 -120.0 -100.0 -80.0 -60.0
Attributes:
units: kelvin
standard_name: air_temperatureWe got the same result, but
we didn’t have to know anything about how the array was created or stored
our code is agnostic about how many dimensions we are dealing with
the intended meaning of our code is much clearer!
Approximate selection and interpolation
With time and space data, we frequently want to sample “near” the coordinate points in our dataset. Here are a few simple ways to achieve that.
Nearest-neighbor sampling
Suppose we want to sample the nearest datapoint within 2 days of date 2018-01-07. Since the last day on our time axis is 2018-01-05, this is well-posed.
.sel has the flexibility to perform nearest neighbor sampling, taking an optional tolerance:
temp.sel(time='2018-01-07', method='nearest', tolerance=timedelta(days=2))
<xarray.DataArray (lat: 3, lon: 4)>
array([[282.15740785, 286.21591172, 279.85464686, 285.96992707],
[290.88477194, 281.37684085, 281.49092035, 282.16621324],
[283.85810564, 285.68428321, 277.78787953, 285.17050016]])
Coordinates:
time datetime64[ns] 2018-01-05
* lat (lat) float64 25.0 40.0 55.0
* lon (lon) float64 -120.0 -100.0 -80.0 -60.0
Attributes:
units: kelvin
standard_name: air_temperaturewhere we see that .sel indeed pulled out the data for date 2018-01-05.
Interpolation
Suppose we want to extract a timeseries for Boulder (40°N, 105°W). Since lon=-105 is not a point on our longitude axis, this requires interpolation between data points.
The .interp() method (see the docs here) works similarly to .sel(). Using .interp(), we can interpolate to any latitude/longitude location:
temp.interp(lon=-105, lat=40)
<xarray.DataArray (time: 5)>
array([283.39098613, 283.25170717, 284.17967965, 278.04106544,
283.75382362])
Coordinates:
* time (time) datetime64[ns] 2018-01-01 2018-01-02 ... 2018-01-05
lon int64 -105
lat int64 40
Attributes:
units: kelvin
standard_name: air_temperatureInfo
Xarray’s interpolation functionality requires the SciPy package!
Slicing along coordinates
Frequently we want to select a range (or slice) along one or more coordinate(s). We can achieve this by passing a Python slice object to .sel(), as follows:
temp.sel(
time=slice('2018-01-01', '2018-01-03'), lon=slice(-110, -70), lat=slice(25, 45)
)
<xarray.DataArray (time: 3, lat: 2, lon: 2)>
array([[[277.95199244, 282.03019204],
[280.71852405, 288.30962373]],
[[285.37426651, 275.88078048],
[280.81104924, 275.86434679]],
[[285.66779544, 275.7531373 ],
[283.67698247, 281.84754872]]])
Coordinates:
* time (time) datetime64[ns] 2018-01-01 2018-01-02 2018-01-03
* lat (lat) float64 25.0 40.0
* lon (lon) float64 -100.0 -80.0
Attributes:
units: kelvin
standard_name: air_temperatureInfo
The calling sequence for slice always looks like slice(start, stop[, step]), where step is optional.
Notice how the length of each coordinate axis has changed due to our slicing.
One more selection method: .loc
All of these operations can also be done within square brackets on the .loc attribute of the DataArray:
temp.loc['2018-01-02']
<xarray.DataArray (lat: 3, lon: 4)>
array([[275.53284611, 285.37426651, 275.88078048, 285.61579451],
[290.57368097, 280.81104924, 275.86434679, 280.78811485],
[278.07454137, 290.51130363, 291.4245341 , 284.55193302]])
Coordinates:
time datetime64[ns] 2018-01-02
* lat (lat) float64 25.0 40.0 55.0
* lon (lon) float64 -120.0 -100.0 -80.0 -60.0
Attributes:
units: kelvin
standard_name: air_temperatureThis is sort of in between the NumPy-style selection
temp[1,:,:]
and the fully label-based selection using .sel()
With .loc, we make use of the coordinate values, but lose the ability to specify the names of the various dimensions. Instead, the slicing must be done in the correct order:
temp.loc['2018-01-01':'2018-01-03', 25:45, -110:-70]
<xarray.DataArray (time: 3, lat: 2, lon: 2)>
array([[[277.95199244, 282.03019204],
[280.71852405, 288.30962373]],
[[285.37426651, 275.88078048],
[280.81104924, 275.86434679]],
[[285.66779544, 275.7531373 ],
[283.67698247, 281.84754872]]])
Coordinates:
* time (time) datetime64[ns] 2018-01-01 2018-01-02 2018-01-03
* lat (lat) float64 25.0 40.0
* lon (lon) float64 -100.0 -80.0
Attributes:
units: kelvin
standard_name: air_temperatureOne advantage of using .loc is that we can use NumPy-style slice notation like 25:45, rather than the more verbose slice(25,45). But of course that also works:
temp.loc['2018-01-01':'2018-01-03', slice(25, 45), -110:-70]
<xarray.DataArray (time: 3, lat: 2, lon: 2)>
array([[[277.95199244, 282.03019204],
[280.71852405, 288.30962373]],
[[285.37426651, 275.88078048],
[280.81104924, 275.86434679]],
[[285.66779544, 275.7531373 ],
[283.67698247, 281.84754872]]])
Coordinates:
* time (time) datetime64[ns] 2018-01-01 2018-01-02 2018-01-03
* lat (lat) float64 25.0 40.0
* lon (lon) float64 -100.0 -80.0
Attributes:
units: kelvin
standard_name: air_temperatureWhat doesn’t work is passing the slices in a different order:
# This will generate an error
# temp.loc[-110:-70, 25:45,'2018-01-01':'2018-01-03']
Opening netCDF data
With its close ties to the netCDF data model, Xarray also supports netCDF as a first-class file format. This means it has easy support for opening netCDF datasets, so long as they conform to some of Xarray’s limitations (such as 1-dimensional coordinates).
Access netCDF data with xr.open_dataset
Once we have a valid path to a data file that Xarray knows how to read, we can open it like this:
ds = xr.open_dataset("../data/comble/radar/anxkazrcfrgeqcM1.b1.20200312.000000.nc").compute()
ds
<xarray.Dataset>
Dimensions: (time: 1737, range: 600, frequency: 1,
sweep: 1, r_calib: 1)
Coordinates:
* time (time) datetime64[ns] 2020-03-12T00:0...
* frequency (frequency) float32 3.489e+10
* range (range) float32 100.7 ... 1.806e+04
azimuth (time) float32 0.0 0.0 0.0 ... 0.0 0.0
elevation (time) float32 90.0 90.0 ... 90.0 90.0
Dimensions without coordinates: sweep, r_calib
Data variables: (12/38)
base_time datetime64[ns] 2020-03-12
time_offset (time) datetime64[ns] 2020-03-12T00:0...
linear_depolarization_ratio (time, range) float32 52.99 ... 19.53
mean_doppler_velocity (time, range) float32 -0.5536 ... -2.209
mean_doppler_velocity_crosspolar_v (time, range) float32 nan nan ... nan
reflectivity (time, range) float32 -52.99 ... -19.53
... ...
longitude float32 15.68
altitude float32 2.0
altitude_agl float32 nan
lat float32 69.14
lon float32 15.68
alt float32 2.0
Attributes: (12/33)
command_line: kazrcfrqc -D 2 -s anx -f M1 -R -n kazrcfrgeqc -...
Conventions: ARM-1.2 CF/Radial-1.4 instrument_parameters rad...
process_version: ingest-kazrcfrqc-0.0-0.dev0.dirty.4.12.14-197.7...
dod_version: kazrcfrgeqc-b1-1.0
input_source: /data/collection/anx/anxkazrM1.00/KAZR_MOMENTS_...
site_id: anx
... ...
scan_name:
software_version: 1.7.6 (Wed Mar 23 17:10:35 UTC 2016 leachman
title: ARM KAZR Moments B1
transform_history: Variable 'censor_mask' set as a bit mask. SNR ...
doi: 10.5439/1478370
history: created by user schuman on machine cirrus16.ccs...Read in Multiple Files Using open_mfdataset
files = sorted(glob.glob("../data/comble/radar/*"))
ds = xr.open_mfdataset(files).compute()
Subsetting the Dataset
Our call to xr.open_dataset() above returned a Dataset object that we’ve decided to call ds. We can then pull out individual fields:
ds.reflectivity
<xarray.DataArray 'reflectivity' (time: 83334, range: 600)>
array([[-52.98858 , -33.44188 , -22.70005 , ..., -20.438488 ,
-18.394522 , -18.08537 ],
[-54.271557 , -37.595207 , -24.503832 , ..., -18.23519 ,
-18.427814 , -18.343391 ],
[-57.0884 , -35.749813 , -23.610859 , ..., -19.081789 ,
-20.370712 , -19.395697 ],
...,
[-54.291916 , -10.5314 , 0.06862259, ..., -19.969387 ,
-16.157347 , -20.416422 ],
[-54.820114 , -10.11786 , 0.45381927, ..., -19.670504 ,
-21.49987 , -20.728188 ],
[-53.631023 , -8.151935 , 1.9688454 , ..., -19.870188 ,
-17.26012 , -21.931446 ]], dtype=float32)
Coordinates:
* time (time) datetime64[ns] 2020-03-12T00:00:00.835386 ... 2020-03-1...
* range (range) float32 100.7 130.7 160.6 ... 1.8e+04 1.803e+04 1.806e+04
azimuth (time) float32 0.0 0.0 0.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0 0.0
elevation (time) float32 90.0 90.0 90.0 90.0 90.0 ... 90.0 90.0 90.0 90.0
Attributes:
long_name: Equivalent reflectivity factor
units: dBZ
standard_name: equivalent_reflectivity_factor
applied_bias_correction: [](recall that we can also use dictionary syntax like ds['isobaric1'] to do the same thing)
Aggregation operations
Not only can you use the named dimensions for manual slicing and indexing of data, but you can also use it to control aggregation operations, like std (standard deviation):
reflectivity = ds['reflectivity']
reflectivity.std(dim=['range'])
<xarray.DataArray 'reflectivity' (time: 83334)>
array([ 9.987671, 10.009455, 9.983448, ..., 12.624017, 12.583106,
12.608554], dtype=float32)
Coordinates:
* time (time) datetime64[ns] 2020-03-12T00:00:00.835386 ... 2020-03-1...
azimuth (time) float32 0.0 0.0 0.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0 0.0
elevation (time) float32 90.0 90.0 90.0 90.0 90.0 ... 90.0 90.0 90.0 90.0Info
Aggregation methods for Xarray objects operate over the named coordinate dimension(s) specified by keyword argument dim. Compare to NumPy, where aggregations operate over specified numbered axes.
Using the sample dataset, we can calculate the temperature profile across our time period!
ref = ds.reflectivity
ref_lowest_5000m = ref.sel(range=slice(0., 5000))
prof = ref_lowest_5000m.mean(dim="time")
prof
<xarray.DataArray 'reflectivity' (range: 164)>
array([-54.97283 , -27.448778 , -20.162565 , -19.562199 , -18.778477 ,
-18.119287 , -17.527794 , -16.988394 , -16.463007 , -15.967142 ,
-15.499426 , -15.051485 , -14.6151285, -14.180205 , -13.741215 ,
-13.314495 , -12.919496 , -12.519125 , -12.125657 , -11.793983 ,
-11.46346 , -11.136843 , -10.837575 , -10.579378 , -10.352728 ,
-10.121803 , -9.91838 , -9.727786 , -9.562 , -9.403213 ,
-9.278362 , -9.164416 , -9.043377 , -8.949946 , -8.87279 ,
-8.785539 , -8.714188 , -8.660834 , -8.63176 , -8.631797 ,
-8.655435 , -8.736296 , -8.838785 , -8.97232 , -9.109644 ,
-9.261291 , -9.415421 , -9.583023 , -9.780044 , -9.9863405,
-10.229948 , -10.498003 , -10.806079 , -11.150559 , -11.517881 ,
-11.883155 , -12.284343 , -12.71636 , -13.150287 , -13.612416 ,
-14.078413 , -14.548593 , -14.97536 , -15.353441 , -15.719428 ,
-16.105438 , -16.462404 , -16.75374 , -17.007557 , -17.256771 ,
-17.526138 , -17.760715 , -18.019426 , -18.311287 , -18.61053 ,
-18.948696 , -19.272732 , -19.578465 , -19.825418 , -19.968918 ,
-20.109495 , -20.227057 , -20.284681 , -20.295177 , -20.303745 ,
-20.319271 , -20.342592 , -20.358269 , -20.396446 , -20.431572 ,
-20.457926 , -20.460478 , -20.448437 , -20.43627 , -20.421385 ,
-20.403185 , -20.37756 , -20.353485 , -20.315321 , -20.319935 ,
-20.34569 , -20.38963 , -20.437658 , -20.493881 , -20.569586 ,
-20.660471 , -20.765326 , -20.883121 , -21.032913 , -21.195692 ,
-21.407215 , -21.640852 , -21.891891 , -22.179762 , -22.432667 ,
-22.704782 , -22.983511 , -23.225466 , -23.498125 , -23.770803 ,
-24.01657 , -24.269527 , -24.522238 , -24.75132 , -24.98275 ,
-25.208271 , -25.422485 , -25.658016 , -25.903503 , -26.134922 ,
-26.398083 , -26.624973 , -26.863201 , -27.099102 , -27.301062 ,
-27.483305 , -27.609858 , -27.728413 , -27.851976 , -27.962738 ,
-28.049582 , -28.13378 , -28.235912 , -28.341232 , -28.436846 ,
-28.519205 , -28.613474 , -28.704872 , -28.763172 , -28.83498 ,
-28.902283 , -28.941643 , -28.963448 , -28.980585 , -29.02627 ,
-29.050957 , -29.09152 , -29.152311 , -29.214195 , -29.281126 ,
-29.355896 , -29.426785 , -29.483341 , -29.53072 ], dtype=float32)
Coordinates:
* range (range) float32 100.7 130.7 160.6 ... 4.927e+03 4.957e+03 4.987e+03Plotting with Xarray
Another major benefit of using labeled data structures is that they enable automated plotting with sensible axis labels.
Simple visualization with .plot()
Much like we saw in Pandas, Xarray includes an interface to Matplotlib that we can access through the .plot() method of every DataArray.
For quick and easy data exploration, we can just call .plot() without any modifiers:
prof.plot();
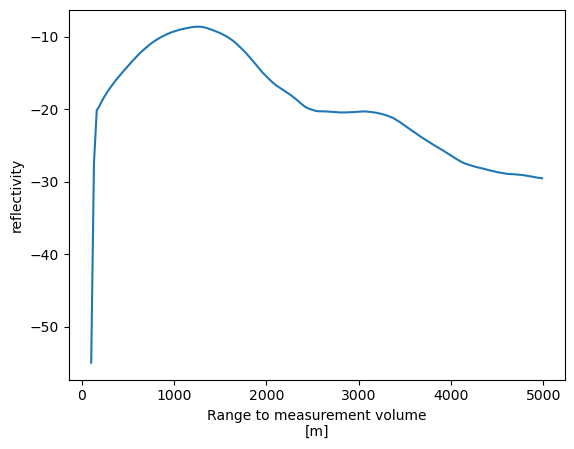
Here Xarray has generated a line plot of the temperature data against the coordinate variable isobaric. Also the metadata are used to auto-generate axis labels and units.
Customizing the plot
As in Pandas, the .plot() method is mostly just a wrapper to Matplotlib, so we can customize our plot in familiar ways.
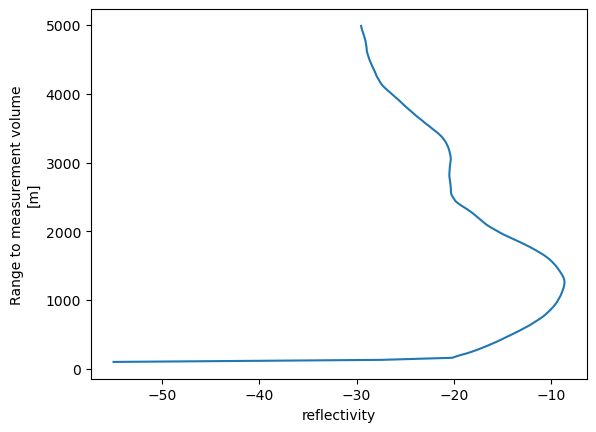
In this air temperature profile example, we would like to make two changes:
swap the axes so that we have isobaric levels on the y (vertical) axis of the figure
make pressure decrease upward in the figure, so that up is up
A few keyword arguments to our .plot() call will take care of this:
prof.plot(y="range")
[<matplotlib.lines.Line2D at 0x29c367d50>]
Plotting 2D data
In the example above, the .plot() method produced a line plot.
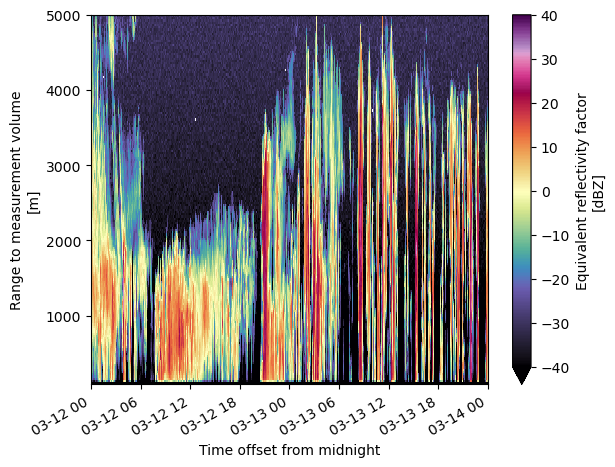
What if we call .plot() on a 2D array?
ref.sel(range=slice(0, 5000)).plot(y='range',
cmap='ChaseSpectral',
vmin=-40,
vmax=40)
<matplotlib.collections.QuadMesh at 0x2a6600590>
We can also make this interactive!
ref.sel(range=slice(0, 5000)).hvplot(x='time',
y='range',
cmap='ChaseSpectral',
clim=(-20, 40),
rasterize=True)
WARNING:param.Image04834: Image dimension time is not evenly sampled to relative tolerance of 0.001. Please use the QuadMesh element for irregularly sampled data or set a higher tolerance on hv.config.image_rtol or the rtol parameter in the Image constructor.
WARNING:param.Image04834: Image dimension time is not evenly sampled to relative tolerance of 0.001. Please use the QuadMesh element for irregularly sampled data or set a higher tolerance on hv.config.image_rtol or the rtol parameter in the Image constructor.
ds.reflectivity.sel(range=slice(0, 5000)).plot(y='range', cmap='Spectral_r');
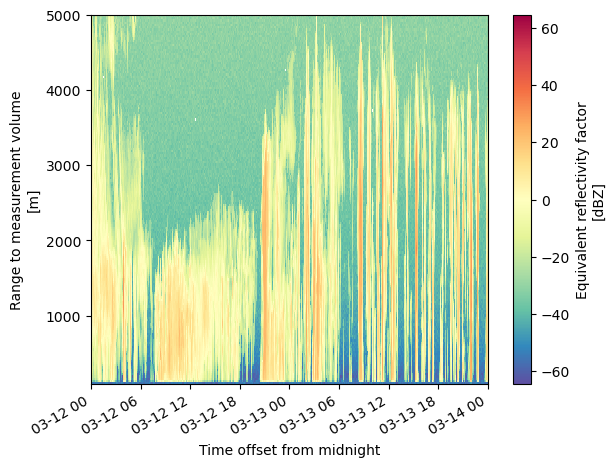
ds.reflectivity.sel(range=slice(0, 5000)).hvplot(x='time', y='range', cmap='Spectral_r', rasterize=True, clabel='Reflectivity (dBZ)')
WARNING:param.Image06073: Image dimension time is not evenly sampled to relative tolerance of 0.001. Please use the QuadMesh element for irregularly sampled data or set a higher tolerance on hv.config.image_rtol or the rtol parameter in the Image constructor.
WARNING:param.Image06073: Image dimension time is not evenly sampled to relative tolerance of 0.001. Please use the QuadMesh element for irregularly sampled data or set a higher tolerance on hv.config.image_rtol or the rtol parameter in the Image constructor.
Customize our Interactive Plots
Our time axis doesn’t tell us much… we can change that! Also note that we add additional parameters to customize our view of the field.
formatter = DatetimeTickFormatter(hours="%d %b %Y \n %H:%M UTC")
reflectivity_plot = ds.reflectivity.sel(range=slice(0, 5000)).hvplot(x='time', y='range', cmap='Spectral_r', xformatter=formatter, clim=(-20, 40), rasterize=True, clabel='Reflectivity (dBZ)')
reflectivity_plot
WARNING:param.Image06893: Image dimension time is not evenly sampled to relative tolerance of 0.001. Please use the QuadMesh element for irregularly sampled data or set a higher tolerance on hv.config.image_rtol or the rtol parameter in the Image constructor.
WARNING:param.Image06893: Image dimension time is not evenly sampled to relative tolerance of 0.001. Please use the QuadMesh element for irregularly sampled data or set a higher tolerance on hv.config.image_rtol or the rtol parameter in the Image constructor.
And the same for velocity…
velocity_plot = ds.mean_doppler_velocity.sel(range=slice(0, 5000)).hvplot(x='time', y='range', cmap='seismic', xformatter=formatter, clim=(-5, 5), rasterize=True, clabel='Mean Doppler Velocity (m/s)')
velocity_plot
WARNING:param.Image07300: Image dimension time is not evenly sampled to relative tolerance of 0.001. Please use the QuadMesh element for irregularly sampled data or set a higher tolerance on hv.config.image_rtol or the rtol parameter in the Image constructor.
WARNING:param.Image07300: Image dimension time is not evenly sampled to relative tolerance of 0.001. Please use the QuadMesh element for irregularly sampled data or set a higher tolerance on hv.config.image_rtol or the rtol parameter in the Image constructor.
Combine our Plots
Now that we have our interactive plots, we can combine them using +
reflectivity_plot + velocity_plot
Or stacked on top of each other…
(reflectivity_plot + velocity_plot).cols(1)
Summary
Xarray brings the joy of Pandas-style labeled data operations to N-dimensional data. As such, it has become a central workhorse in the geoscience community for the analysis of gridded datasets. Xarray allows us to open self-describing NetCDF files and make full use of the coordinate axes, labels, units, and other metadata. By making use of labeled coordinates, our code is often easier to write, easier to read, and more robust.
We also covered some interactive plots using xarray and hvPlot!
What’s next?
Additional notebooks to appear in this section will go into more detail about
arithemtic and broadcasting with Xarray data structures
using “group by” operations
remote data access with OpenDAP
more advanced visualization including map integration with Cartopy
Resources and references
This notebook was adapated from material in Unidata’s Python Training.
The best resource for Xarray is the Xarray documentation. See in particular
Another excellent resource is this Xarray Tutorial collection.

