
Computations and Masks with Xarray#
Overview#
In this notebook, we will:
Do basic arithmetic with DataArrays and Datasets
Perform aggregation (reduction) along one or multiple dimensions of a DataArray or Dataset
Compute climatology and anomaly using xarray’s “split-apply-combine” approach via
.groupby()Perform weighted reductions along one or multiple dimensions of a DataArray or Dataset
Provide an overview of masking data in xarray
Mask data using
.where()method
Prerequisites#
Concepts |
Importance |
Notes |
|---|---|---|
Necessary |
Time to learn: 30 minutes
Imports#
import matplotlib.pyplot as plt
import numpy as np
import xarray as xr
Let’s open the monthly sea surface temperature (SST) data from the Community Earth System Model v2 (CESM2), which is a Global Climate Model:
ds = xr.open_dataset("data/CESM2_sst_data.nc")
ds
<xarray.Dataset>
Dimensions: (time: 180, d2: 2, lat: 180, lon: 360)
Coordinates:
* time (time) object 2000-01-15 12:00:00 ... 2014-12-15 12:00:00
* lat (lat) float64 -89.5 -88.5 -87.5 -86.5 ... 86.5 87.5 88.5 89.5
* lon (lon) float64 0.5 1.5 2.5 3.5 4.5 ... 356.5 357.5 358.5 359.5
Dimensions without coordinates: d2
Data variables:
time_bnds (time, d2) object 2000-01-01 00:00:00 ... 2015-01-01 00:00:00
lat_bnds (lat, d2) float64 -90.0 -89.0 -89.0 -88.0 ... 88.0 89.0 89.0 90.0
lon_bnds (lon, d2) float64 0.0 1.0 1.0 2.0 2.0 ... 358.0 359.0 359.0 360.0
tos (time, lat, lon) float32 ...
Attributes: (12/45)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 674885.0
branch_time_in_parent: 219000.0
case_id: 972
... ...
sub_experiment_id: none
table_id: Omon
tracking_id: hdl:21.14100/2975ffd3-1d7b-47e3-961a-33f212ea4eb2
variable_id: tos
variant_info: CMIP6 20th century experiments (1850-2014) with C...
variant_label: r11i1p1f1Convert our cftime to datetime#
You’ll notice that by default, we have times in cftime - let’s convert those back to regular datetime!
ds["time"] = ds.indexes["time"].to_datetimeindex()
/var/folders/dd/zgpcpccj46q2tfrvpwpp3r5w0000gn/T/ipykernel_7911/3354600616.py:1: RuntimeWarning: Converting a CFTimeIndex with dates from a non-standard calendar, 'noleap', to a pandas.DatetimeIndex, which uses dates from the standard calendar. This may lead to subtle errors in operations that depend on the length of time between dates.
ds["time"] = ds.indexes["time"].to_datetimeindex()
Arithmetic Operations#
Arithmetic operations with a single DataArray automatically apply over all array values (like NumPy). This process is called vectorization. Let’s convert the air temperature from degrees Celsius to kelvins:
ds.tos + 273.15
<xarray.DataArray 'tos' (time: 180, lat: 180, lon: 360)>
array([[[ nan, nan, nan, ..., nan, nan,
nan],
[ nan, nan, nan, ..., nan, nan,
nan],
[ nan, nan, nan, ..., nan, nan,
nan],
...,
[271.3552 , 271.3553 , 271.3554 , ..., 271.35495, 271.355 ,
271.3551 ],
[271.36005, 271.36014, 271.36023, ..., 271.35986, 271.35992,
271.36 ],
[271.36447, 271.36453, 271.3646 , ..., 271.3643 , 271.36435,
271.3644 ]],
[[ nan, nan, nan, ..., nan, nan,
nan],
[ nan, nan, nan, ..., nan, nan,
nan],
[ nan, nan, nan, ..., nan, nan,
nan],
...
[271.40677, 271.40674, 271.4067 , ..., 271.40695, 271.4069 ,
271.40683],
[271.41296, 271.41293, 271.41293, ..., 271.41306, 271.413 ,
271.41296],
[271.41772, 271.41772, 271.41772, ..., 271.41766, 271.4177 ,
271.4177 ]],
[[ nan, nan, nan, ..., nan, nan,
nan],
[ nan, nan, nan, ..., nan, nan,
nan],
[ nan, nan, nan, ..., nan, nan,
nan],
...,
[271.39386, 271.39383, 271.3938 , ..., 271.39407, 271.394 ,
271.39392],
[271.39935, 271.39932, 271.39932, ..., 271.39948, 271.39944,
271.39938],
[271.40372, 271.40372, 271.40375, ..., 271.4037 , 271.4037 ,
271.40372]]], dtype=float32)
Coordinates:
* time (time) datetime64[ns] 2000-01-15T12:00:00 ... 2014-12-15T12:00:00
* lat (lat) float64 -89.5 -88.5 -87.5 -86.5 -85.5 ... 86.5 87.5 88.5 89.5
* lon (lon) float64 0.5 1.5 2.5 3.5 4.5 ... 355.5 356.5 357.5 358.5 359.5Lets’s square all values in tos:
ds.tos**2
<xarray.DataArray 'tos' (time: 180, lat: 180, lon: 360)>
array([[[ nan, nan, nan, ..., nan, nan,
nan],
[ nan, nan, nan, ..., nan, nan,
nan],
[ nan, nan, nan, ..., nan, nan,
nan],
...,
[3.2213385, 3.2209656, 3.220537 , ..., 3.2221622, 3.221913 ,
3.2216525],
[3.203904 , 3.203617 , 3.2032912, ..., 3.2045207, 3.2043478,
3.2041442],
[3.1881146, 3.1879027, 3.1876712, ..., 3.188714 , 3.1885312,
3.1883302]],
[[ nan, nan, nan, ..., nan, nan,
nan],
[ nan, nan, nan, ..., nan, nan,
nan],
[ nan, nan, nan, ..., nan, nan,
nan],
...
[3.0388296, 3.0389647, 3.0390673, ..., 3.038165 , 3.0383828,
3.0386322],
[3.0173173, 3.0173445, 3.0173297, ..., 3.0169601, 3.0171173,
3.0172386],
[3.000791 , 3.0007784, 3.0007539, ..., 3.000933 , 3.000896 ,
3.0008452]],
[[ nan, nan, nan, ..., nan, nan,
nan],
[ nan, nan, nan, ..., nan, nan,
nan],
[ nan, nan, nan, ..., nan, nan,
nan],
...,
[3.0839543, 3.0841148, 3.0842566, ..., 3.0832636, 3.0834875,
3.0837412],
[3.064733 , 3.0648024, 3.0648358, ..., 3.0642793, 3.0644639,
3.0646174],
[3.0494578, 3.0494475, 3.0494263, ..., 3.049596 , 3.0495603,
3.0495107]]], dtype=float32)
Coordinates:
* time (time) datetime64[ns] 2000-01-15T12:00:00 ... 2014-12-15T12:00:00
* lat (lat) float64 -89.5 -88.5 -87.5 -86.5 -85.5 ... 86.5 87.5 88.5 89.5
* lon (lon) float64 0.5 1.5 2.5 3.5 4.5 ... 355.5 356.5 357.5 358.5 359.5Aggregation Methods#
A very common step during data analysis is to summarize the data in question by computing aggregations like sum(), mean(), median(), min(), max() in which reduced data provide insight into the nature of large dataset. Let’s explore some of these aggregation methods.
Compute the mean:
ds.tos.mean()
<xarray.DataArray 'tos' ()> array(14.250171, dtype=float32)
Because we specified no dim argument the function was applied over all dimensions, computing the mean of every element of tos across time and space. It is possible to specify a dimension along which to compute an aggregation. For example, to calculate the mean in time for all locations, specify the time dimension as the dimension along which the mean should be calculated:
ds.tos.mean(dim='time').plot(size=7);
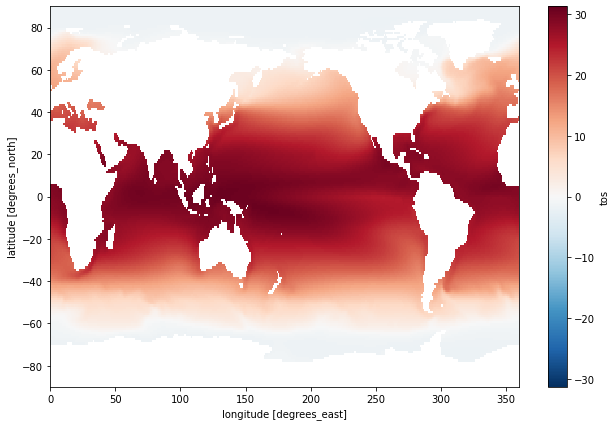
Compute the temporal min:
ds.tos.min(dim=['time'])
<xarray.DataArray 'tos' (lat: 180, lon: 360)>
array([[ nan, nan, nan, ..., nan, nan,
nan],
[ nan, nan, nan, ..., nan, nan,
nan],
[ nan, nan, nan, ..., nan, nan,
nan],
...,
[-1.8083605, -1.8083031, -1.8082187, ..., -1.8083988, -1.8083944,
-1.8083915],
[-1.8025414, -1.8024837, -1.8024155, ..., -1.8026428, -1.8026177,
-1.8025846],
[-1.7984415, -1.7983989, -1.7983514, ..., -1.7985678, -1.7985296,
-1.7984871]], dtype=float32)
Coordinates:
* lat (lat) float64 -89.5 -88.5 -87.5 -86.5 -85.5 ... 86.5 87.5 88.5 89.5
* lon (lon) float64 0.5 1.5 2.5 3.5 4.5 ... 355.5 356.5 357.5 358.5 359.5Compute the spatial sum:
ds.tos.sum(dim=['lat', 'lon'])
<xarray.DataArray 'tos' (time: 180)>
array([603767. , 607702.5 , 603976.5 , 599373.56, 595119.94, 595716.75,
598177.3 , 600670.6 , 597825.56, 591869. , 590507.7 , 597189.2 ,
605954.06, 609151. , 606868.9 , 602329.9 , 599465.75, 601205.5 ,
605144.4 , 608588.5 , 604046.9 , 598927.75, 597519.75, 603876.9 ,
612424.44, 615765.2 , 612615.44, 606310.6 , 602034.4 , 600784.9 ,
602013.5 , 603142.2 , 598850.9 , 591917.44, 589234.56, 596162.5 ,
602942.06, 607196.9 , 604928.2 , 601735.6 , 599011.8 , 599490.9 ,
600801.44, 602786.94, 598867.2 , 594081.8 , 593736.25, 598995.6 ,
607285.25, 611901.06, 609562.75, 603527.3 , 600215.4 , 601372.6 ,
604144.5 , 605376.75, 601256.2 , 595245.2 , 594002.06, 600490.4 ,
611878.6 , 616563. , 613050.8 , 605734. , 600808.75, 600898.06,
603930.56, 605644.7 , 599917.5 , 592048.06, 590082.8 , 596950.7 ,
607701.94, 610844.7 , 609509.6 , 603380.94, 599838.1 , 600334.25,
604386.6 , 607848.1 , 602155.2 , 594949.06, 593815.06, 598365.3 ,
608730.8 , 612056.5 , 609922.5 , 603077.1 , 600134.1 , 602821.2 ,
606152.75, 610257.8 , 604685.8 , 596858. , 592894.8 , 599944.9 ,
609764.44, 614610.75, 611434.75, 605606.4 , 603790.94, 605750.2 ,
609250.06, 612935.7 , 609645.06, 601706.4 , 598896.5 , 605349.75,
614671.8 , 618686.7 , 615895.2 , 609438.2 , 605399.56, 606126.75,
607942.3 , 609680.4 , 604814.25, 595841.94, 591908.44, 595638.7 ,
604798.94, 611327.1 , 609765.7 , 603727.56, 600970. , 602514. ,
606303.7 , 609225.25, 603724.3 , 595944.8 , 594477.4 , 597807.4 ,
607379.06, 611808.56, 610112.94, 607196.3 , 604733.06, 605488.25,
610048.3 , 612655.75, 608906.25, 602349.7 , 601754.2 , 609220.4 ,
619367.1 , 623783.2 , 619949.7 , 613369.06, 610190.8 , 611091.2 ,
614213.44, 615665.06, 611722.2 , 606259.56, 605970.2 , 611463.3 ,
619794.6 , 626036.5 , 623085.44, 616295.9 , 611886.3 , 611881.9 ,
614420.75, 616853.56, 610375.44, 603471.5 , 602108.25, 608094.3 ,
617450.7 , 623508.7 , 619830.2 , 612033.3 , 608737.2 , 610105.25,
613692.7 , 616360.44, 611735.4 , 606512.7 , 604249.44, 608777.44],
dtype=float32)
Coordinates:
* time (time) datetime64[ns] 2000-01-15T12:00:00 ... 2014-12-15T12:00:00Compute the temporal median:
ds.tos.median(dim='time')
<xarray.DataArray 'tos' (lat: 180, lon: 360)>
array([[ nan, nan, nan, ..., nan, nan,
nan],
[ nan, nan, nan, ..., nan, nan,
nan],
[ nan, nan, nan, ..., nan, nan,
nan],
...,
[-1.7648907, -1.7648032, -1.7647004, ..., -1.7650614, -1.7650102,
-1.7649589],
[-1.7590305, -1.7589546, -1.7588665, ..., -1.7591925, -1.7591486,
-1.759095 ],
[-1.7536805, -1.753602 , -1.7535168, ..., -1.753901 , -1.753833 ,
-1.7537591]], dtype=float32)
Coordinates:
* lat (lat) float64 -89.5 -88.5 -87.5 -86.5 -85.5 ... 86.5 87.5 88.5 89.5
* lon (lon) float64 0.5 1.5 2.5 3.5 4.5 ... 355.5 356.5 357.5 358.5 359.5The following table summarizes some other built-in xarray aggregations:
Aggregation |
Description |
|---|---|
|
Total number of items |
|
Mean and median |
|
Minimum and maximum |
|
Standard deviation and variance |
|
Compute product of elements |
|
Compute sum of elements |
|
Find index of minimum and maximum value |
GroupBy: Split, Apply, Combine#
Simple aggregations can give useful summary of our dataset, but often we would prefer to aggregate conditionally on some coordinate labels or groups. Xarray provides the so-called groupby operation which enables the split-apply-combine workflow on xarray DataArrays and Datasets. The split-apply-combine operation is illustrated in this figure

This makes clear what the groupby accomplishes:
The split step involves breaking up and grouping an xarray Dataset or DataArray depending on the value of the specified group key.
The apply step involves computing some function, usually an aggregate, transformation, or filtering, within the individual groups.
The combine step merges the results of these operations into an output xarray Dataset or DataArray.
We are going to use groupby to remove the seasonal cycle (“climatology”) from our dataset. See the xarray groupby user guide for more examples of what groupby can take as an input.
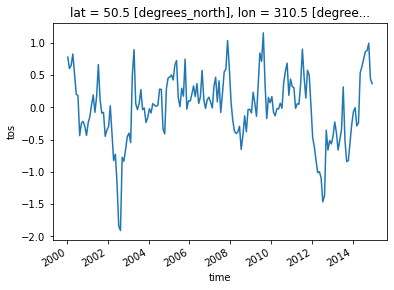
First, let’s select a gridpoint closest to a specified lat-lon, and plot a time series of SST at that point. The annual cycle will be quite evident.
ds.tos.sel(lon=310, lat=50, method='nearest').plot();
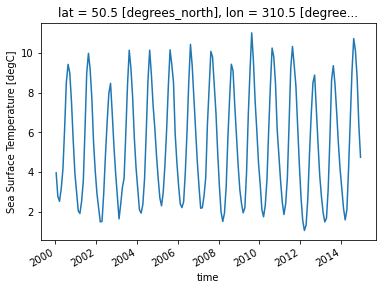
Split#
Let’s group data by month, i.e. all Januaries in one group, all Februaries in one group, etc.
ds.tos.groupby(ds.time.dt.month)
DataArrayGroupBy, grouped over 'month'
12 groups with labels 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12.
In the above code, we are using the .dt DatetimeAccessor to extract specific components of dates/times in our time coordinate dimension. For example, we can extract the year with ds.time.dt.year. See also the equivalent Pandas documentation.
Xarray also offers a more concise syntax when the variable you’re grouping on is already present in the dataset. This is identical to ds.tos.groupby(ds.time.dt.month):
ds.tos.groupby('time.month')
DataArrayGroupBy, grouped over 'month'
12 groups with labels 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12.
Apply & Combine#
Now that we have groups defined, it’s time to “apply” a calculation to the group. These calculations can either be:
aggregation: reduces the size of the group
transformation: preserves the group’s full size
At then end of the apply step, xarray will automatically combine the aggregated/transformed groups back into a single object.
Compute climatology#
Let’s calculate the climatology at every point in the dataset:
tos_clim = ds.tos.groupby('time.month').mean()
tos_clim
<xarray.DataArray 'tos' (month: 12, lat: 180, lon: 360)>
array([[[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
...,
[-1.780786 , -1.780688 , -1.7805718, ..., -1.7809757,
-1.7809197, -1.7808627],
[-1.7745041, -1.7744204, -1.7743237, ..., -1.77467 ,
-1.774626 , -1.7745715],
[-1.7691481, -1.7690798, -1.7690051, ..., -1.7693441,
-1.7692844, -1.7692182]],
[[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
...
[-1.7605033, -1.760397 , -1.7602725, ..., -1.760718 ,
-1.7606541, -1.7605885],
[-1.7544289, -1.7543424, -1.7542422, ..., -1.754608 ,
-1.754559 , -1.7545002],
[-1.7492163, -1.749148 , -1.7490736, ..., -1.7494118,
-1.7493519, -1.7492864]],
[[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
...,
[-1.7711828, -1.7710832, -1.7709653, ..., -1.7713748,
-1.7713183, -1.7712607],
[-1.7648666, -1.7647841, -1.7646879, ..., -1.7650299,
-1.7649865, -1.7649331],
[-1.759478 , -1.7594113, -1.7593384, ..., -1.7596704,
-1.7596117, -1.759547 ]]], dtype=float32)
Coordinates:
* lat (lat) float64 -89.5 -88.5 -87.5 -86.5 -85.5 ... 86.5 87.5 88.5 89.5
* lon (lon) float64 0.5 1.5 2.5 3.5 4.5 ... 355.5 356.5 357.5 358.5 359.5
* month (month) int64 1 2 3 4 5 6 7 8 9 10 11 12Plot climatology at a specific point:
tos_clim.sel(lon=310, lat=50, method='nearest').plot();
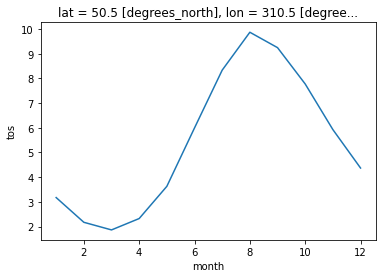
Plot zonal mean climatology:
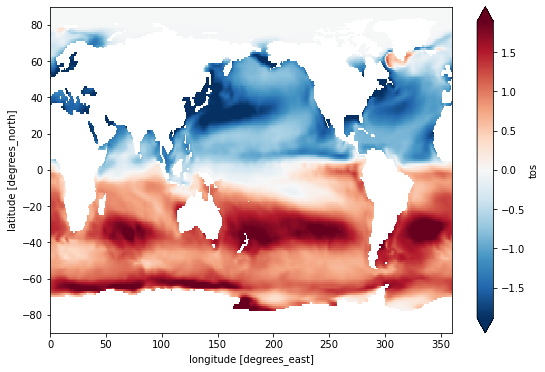
tos_clim.mean(dim='lon').transpose().plot.contourf(levels=12, cmap='turbo');
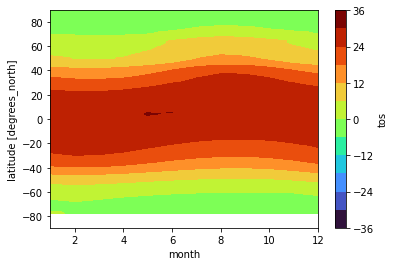
Calculate and plot the difference between January and December climatologies:
(tos_clim.sel(month=1) - tos_clim.sel(month=12)).plot(size=6, robust=True);
Compute anomaly#
Now let’s combine the previous steps to compute climatology and use xarray’s groupby arithmetic to remove this climatology from our original data:
gb = ds.tos.groupby('time.month')
tos_anom = gb - gb.mean(dim='time')
tos_anom
<xarray.DataArray 'tos' (time: 180, lat: 180, lon: 360)>
array([[[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
...,
[-0.01402271, -0.01401687, -0.01401365, ..., -0.01406252,
-0.01404917, -0.01403356],
[-0.01544118, -0.01544476, -0.01545036, ..., -0.0154475 ,
-0.01544321, -0.01544082],
[-0.01638114, -0.01639009, -0.01639998, ..., -0.01635301,
-0.01636147, -0.01637137]],
[[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
...
[ 0.01727939, 0.01713431, 0.01698041, ..., 0.0176847 ,
0.01755834, 0.01742125],
[ 0.0173862 , 0.0172919 , 0.01719594, ..., 0.01766813,
0.01757395, 0.01748013],
[ 0.01693714, 0.01687253, 0.01680517, ..., 0.01709175,
0.0170424 , 0.01699162]],
[[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
...,
[ 0.01506364, 0.01491845, 0.01476014, ..., 0.01545238,
0.0153321 , 0.01520228],
[ 0.0142287 , 0.01412642, 0.01402068, ..., 0.0145216 ,
0.01442552, 0.01432824],
[ 0.01320827, 0.01314461, 0.01307774, ..., 0.0133611 ,
0.0133127 , 0.01326215]]], dtype=float32)
Coordinates:
* time (time) datetime64[ns] 2000-01-15T12:00:00 ... 2014-12-15T12:00:00
* lat (lat) float64 -89.5 -88.5 -87.5 -86.5 -85.5 ... 86.5 87.5 88.5 89.5
* lon (lon) float64 0.5 1.5 2.5 3.5 4.5 ... 355.5 356.5 357.5 358.5 359.5
month (time) int64 1 2 3 4 5 6 7 8 9 10 11 ... 2 3 4 5 6 7 8 9 10 11 12tos_anom.sel(lon=310, lat=50, method='nearest').plot();
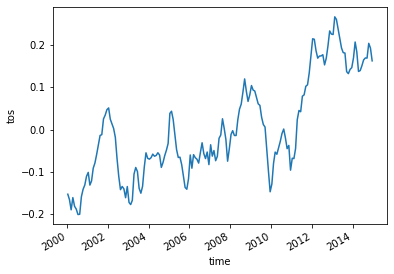
Let’s compute and visualize the mean global anomaly over time. We need to specify both lat and lon dimensions in the dim argument to mean():
unweighted_mean_global_anom = tos_anom.mean(dim=['lat', 'lon'])
unweighted_mean_global_anom.plot();
An operation which combines grid cells of different size is not scientifically valid unless each cell is weighted by the size of the grid cell. Xarray has a convenient .weighted() method to accomplish this
Let’s first load the cell area data from another CESM2 dataset that contains the weights for the grid cells:
areacello = xr.open_dataset("data/CESM2_grid_variables.nc").areacello
areacello
<xarray.DataArray 'areacello' (lat: 180, lon: 360)>
array([[1.078982e+08, 1.078982e+08, 1.078982e+08, ..., 1.078982e+08,
1.078982e+08, 1.078982e+08],
[3.236618e+08, 3.236618e+08, 3.236618e+08, ..., 3.236618e+08,
3.236618e+08, 3.236618e+08],
[5.393268e+08, 5.393268e+08, 5.393268e+08, ..., 5.393268e+08,
5.393268e+08, 5.393268e+08],
...,
[5.393268e+08, 5.393268e+08, 5.393268e+08, ..., 5.393268e+08,
5.393268e+08, 5.393268e+08],
[3.236618e+08, 3.236618e+08, 3.236618e+08, ..., 3.236618e+08,
3.236618e+08, 3.236618e+08],
[1.078982e+08, 1.078982e+08, 1.078982e+08, ..., 1.078982e+08,
1.078982e+08, 1.078982e+08]])
Coordinates:
* lat (lat) float64 -89.5 -88.5 -87.5 -86.5 -85.5 ... 86.5 87.5 88.5 89.5
* lon (lon) float64 0.5 1.5 2.5 3.5 4.5 ... 355.5 356.5 357.5 358.5 359.5
Attributes:
cell_methods: area: sum
comment: TAREA
description: Cell areas for any grid used to report ocean variables an...
frequency: fx
id: areacello
long_name: Grid-Cell Area for Ocean Variables
mipTable: Ofx
out_name: areacello
prov: Ofx ((isd.003))
realm: ocean
standard_name: cell_area
time_label: None
time_title: No temporal dimensions ... fixed field
title: Grid-Cell Area for Ocean Variables
type: real
units: m2
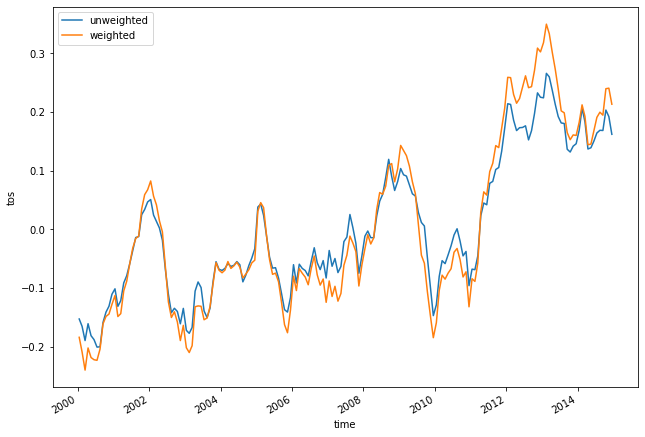
variable_id: areacelloAs before, let’s calculate area-weighted mean global anomaly:
weighted_mean_global_anom = tos_anom.weighted(areacello).mean(dim=['lat', 'lon'])
Let’s plot both unweighted and weighted means:
unweighted_mean_global_anom.plot(size=7)
weighted_mean_global_anom.plot()
plt.legend(['unweighted', 'weighted']);
Other high level computation functionality#
rolling: Useful for computing aggregations on moving windows of your dataset e.g. computing moving averages
For example, resample to annual frequency:
r = ds.tos.resample(time='AS')
r
DataArrayResample, grouped over '__resample_dim__'
15 groups with labels 2000-01-01, ..., 2014-01-01.
r.mean()
<xarray.DataArray 'tos' (time: 15, lat: 180, lon: 360)>
array([[[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
...,
[-1.7474419, -1.7474264, -1.7474008, ..., -1.7474308,
-1.7474365, -1.7474445],
[-1.7424874, -1.7424612, -1.7424251, ..., -1.742536 ,
-1.7425283, -1.7425116],
[-1.7382039, -1.7381679, -1.7381277, ..., -1.7383199,
-1.7382846, -1.7382454]],
[[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
...
[-1.6902231, -1.6899008, -1.6895409, ..., -1.6910189,
-1.6907759, -1.6905178],
[-1.6879102, -1.6876906, -1.6874666, ..., -1.6885366,
-1.6883289, -1.688121 ],
[-1.6883243, -1.6881752, -1.6880217, ..., -1.6886654,
-1.6885542, -1.6884427]],
[[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
...,
[-1.6893266, -1.6893964, -1.6894479, ..., -1.6889572,
-1.6890831, -1.6892204],
[-1.6776317, -1.6777302, -1.6778082, ..., -1.6771463,
-1.6773272, -1.677492 ],
[-1.672563 , -1.6726688, -1.6727766, ..., -1.6723493,
-1.6724195, -1.6724887]]], dtype=float32)
Coordinates:
* time (time) datetime64[ns] 2000-01-01 2001-01-01 ... 2014-01-01
* lat (lat) float64 -89.5 -88.5 -87.5 -86.5 -85.5 ... 86.5 87.5 88.5 89.5
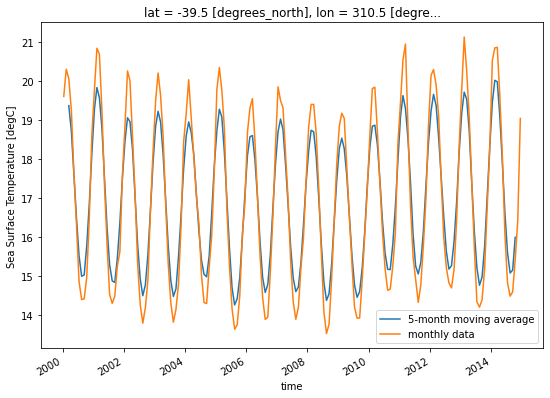
* lon (lon) float64 0.5 1.5 2.5 3.5 4.5 ... 355.5 356.5 357.5 358.5 359.5Compute a 5-month moving average:
m_avg = ds.tos.rolling(time=5, center=True).mean()
m_avg
<xarray.DataArray 'tos' (time: 180, lat: 180, lon: 360)>
array([[[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
...,
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan]],
[[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
...
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan]],
[[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
...,
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan]]])
Coordinates:
* time (time) datetime64[ns] 2000-01-15T12:00:00 ... 2014-12-15T12:00:00
* lat (lat) float64 -89.5 -88.5 -87.5 -86.5 -85.5 ... 86.5 87.5 88.5 89.5
* lon (lon) float64 0.5 1.5 2.5 3.5 4.5 ... 355.5 356.5 357.5 358.5 359.5
Attributes:
cell_measures: area: areacello
cell_methods: area: mean where sea time: mean
comment: Model data on the 1x1 grid includes values in all cells f...
description: This may differ from "surface temperature" in regions of ...
frequency: mon
id: tos
long_name: Sea Surface Temperature
mipTable: Omon
out_name: tos
prov: Omon ((isd.003))
realm: ocean
standard_name: sea_surface_temperature
time: time
time_label: time-mean
time_title: Temporal mean
title: Sea Surface Temperature
type: real
units: degC
variable_id: toslat = 50
lon = 310
m_avg.isel(lat=lat, lon=lon).plot(size=6)
ds.tos.isel(lat=lat, lon=lon).plot()
plt.legend(['5-month moving average', 'monthly data']);
Masking Data#
Using the xr.where() or .where() method, elements of an xarray Dataset or xarray DataArray that satisfy a given condition or multiple conditions can be replaced/masked. To demonstrate this, we are going to use the .where() method on the tos DataArray.
We will use the same sea surface temperature dataset:
ds
<xarray.Dataset>
Dimensions: (time: 180, d2: 2, lat: 180, lon: 360)
Coordinates:
* time (time) datetime64[ns] 2000-01-15T12:00:00 ... 2014-12-15T12:00:00
* lat (lat) float64 -89.5 -88.5 -87.5 -86.5 ... 86.5 87.5 88.5 89.5
* lon (lon) float64 0.5 1.5 2.5 3.5 4.5 ... 356.5 357.5 358.5 359.5
Dimensions without coordinates: d2
Data variables:
time_bnds (time, d2) object 2000-01-01 00:00:00 ... 2015-01-01 00:00:00
lat_bnds (lat, d2) float64 -90.0 -89.0 -89.0 -88.0 ... 88.0 89.0 89.0 90.0
lon_bnds (lon, d2) float64 0.0 1.0 1.0 2.0 2.0 ... 358.0 359.0 359.0 360.0
tos (time, lat, lon) float32 nan nan nan nan ... -1.746 -1.746 -1.746
Attributes: (12/45)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 674885.0
branch_time_in_parent: 219000.0
case_id: 972
... ...
sub_experiment_id: none
table_id: Omon
tracking_id: hdl:21.14100/2975ffd3-1d7b-47e3-961a-33f212ea4eb2
variable_id: tos
variant_info: CMIP6 20th century experiments (1850-2014) with C...
variant_label: r11i1p1f1Using where with one condition#
Imagine we wish to analyze just the last time in the dataset. We could of course use .isel() for this:
sample = ds.tos.isel(time=-1)
sample
<xarray.DataArray 'tos' (lat: 180, lon: 360)>
array([[ nan, nan, nan, ..., nan, nan, nan],
[ nan, nan, nan, ..., nan, nan, nan],
[ nan, nan, nan, ..., nan, nan, nan],
...,
[-1.756119, -1.756165, -1.756205, ..., -1.755922, -1.755986, -1.756058],
[-1.750638, -1.750658, -1.750667, ..., -1.750508, -1.750561, -1.750605],
[-1.74627 , -1.746267, -1.746261, ..., -1.746309, -1.746299, -1.746285]],
dtype=float32)
Coordinates:
time datetime64[ns] 2014-12-15T12:00:00
* lat (lat) float64 -89.5 -88.5 -87.5 -86.5 -85.5 ... 86.5 87.5 88.5 89.5
* lon (lon) float64 0.5 1.5 2.5 3.5 4.5 ... 355.5 356.5 357.5 358.5 359.5
Attributes:
cell_measures: area: areacello
cell_methods: area: mean where sea time: mean
comment: Model data on the 1x1 grid includes values in all cells f...
description: This may differ from "surface temperature" in regions of ...
frequency: mon
id: tos
long_name: Sea Surface Temperature
mipTable: Omon
out_name: tos
prov: Omon ((isd.003))
realm: ocean
standard_name: sea_surface_temperature
time: time
time_label: time-mean
time_title: Temporal mean
title: Sea Surface Temperature
type: real
units: degC
variable_id: tosUnlike .isel() and .sel() that change the shape of the returned results, .where() preserves the shape of the original data. It accomplishes this by returning values from the original DataArray or Dataset if the condition is True, and fills in values (by default nan) wherever the condition is False.
Before applying it, let’s look at the .where() documentation. As the documention points out, the conditional expression in .where() can be:
a DataArray
a Dataset
a function
For demonstration purposes, let’s use .where() to mask locations with temperature values greater than 0:
masked_sample = sample.where(sample < 0.0)
masked_sample
<xarray.DataArray 'tos' (lat: 180, lon: 360)>
array([[ nan, nan, nan, ..., nan, nan,
nan],
[ nan, nan, nan, ..., nan, nan,
nan],
[ nan, nan, nan, ..., nan, nan,
nan],
...,
[-1.7561191, -1.7561648, -1.7562052, ..., -1.7559224, -1.7559862,
-1.7560585],
[-1.7506379, -1.7506577, -1.7506672, ..., -1.7505083, -1.750561 ,
-1.7506049],
[-1.7462697, -1.7462667, -1.7462606, ..., -1.7463093, -1.746299 ,
-1.7462848]], dtype=float32)
Coordinates:
time datetime64[ns] 2014-12-15T12:00:00
* lat (lat) float64 -89.5 -88.5 -87.5 -86.5 -85.5 ... 86.5 87.5 88.5 89.5
* lon (lon) float64 0.5 1.5 2.5 3.5 4.5 ... 355.5 356.5 357.5 358.5 359.5
Attributes:
cell_measures: area: areacello
cell_methods: area: mean where sea time: mean
comment: Model data on the 1x1 grid includes values in all cells f...
description: This may differ from "surface temperature" in regions of ...
frequency: mon
id: tos
long_name: Sea Surface Temperature
mipTable: Omon
out_name: tos
prov: Omon ((isd.003))
realm: ocean
standard_name: sea_surface_temperature
time: time
time_label: time-mean
time_title: Temporal mean
title: Sea Surface Temperature
type: real
units: degC
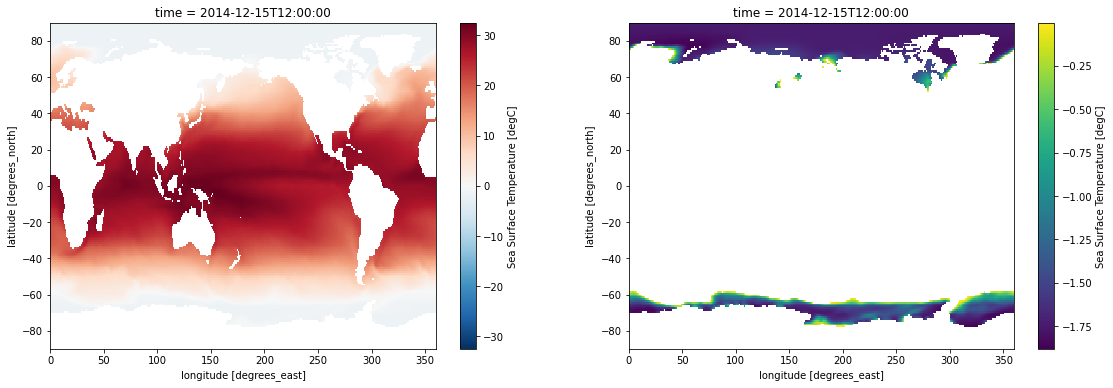
variable_id: tosLet’s plot both our original sample, and the masked sample:
fig, axes = plt.subplots(ncols=2, figsize=(19, 6))
sample.plot(ax=axes[0])
masked_sample.plot(ax=axes[1]);
Using where with multiple conditions#
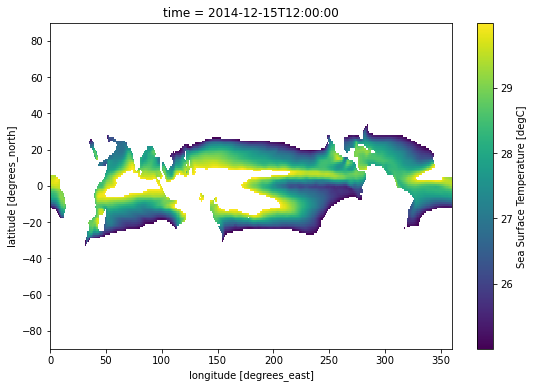
.where() allows providing multiple conditions. To do this, we need to make sure each conditional expression is enclosed in (). To combine conditions, we use the bit-wise and (&) operator and/or the bit-wise or (|). Let’s use .where() to mask locations with temperature values less than 25 and greater than 30:
sample.where((sample > 25) & (sample < 30)).plot(size=6);
We can use coordinates to apply a mask as well. Below, we use the latitude and longitude coordinates to mask everywhere outside of the Niño 3.4 region:
sample.where(
(sample.lat < 5) & (sample.lat > -5) & (sample.lon > 190) & (sample.lon < 240)
).plot(size=6);

Using where with a custom fill value#
.where() can take a second argument, which, if supplied, defines a fill value for the masked region. Below we fill masked regions with a constant 0:
sample.where((sample > 25) & (sample < 30), 0).plot(size=6);
Summary#
Similar to NumPy, arithmetic operations are vectorized over a DataArray
Xarray provides aggregation methods like
sum()andmean(), with the option to specify which dimension over which the operation will be donegroupbyenables the convenient split-apply-combine workflowThe
.where()method allows for filtering or replacing of data based on one or more provided conditions
What’s next?#
In the next notebook, we will work through an example of plotting the Niño 3.4 Index.

