Using EMC² to Process E3SM Output#
Overview of EMC²#
EMC² can be used to simulate radar and lidar observables faithful to large-scale model physics. Here, we will demonstrate using EMC² to evaluate a climate model’s output. We will process E3SM regional output around the fixed ARM site at Utqiagvik, North Slope of Alaska site, using EMC² radiation approach (fastest processing; faithful to E3SM’s radiation logic; similar general approach to that used for the on-line COSP).
We will briefly explore:#
Reading model output file using EMC²
Generating subcolumns and running the radar and lidar instrument simualtors
Classifying the simulator output
Plotting the simulated observables to evaluate the model output
Imports#
import emc2
import matplotlib as mpl
import numpy as np
import xarray as xr
Initializing Instrument Class Objects#
We begin with generating instrument class objects representing instruments deployed at Utqiagvik; that is, high spectral resolution lidar (HSRL) emc2.core.instruments.HSRL and ARM Ka-band Zenith Radar (KAZR) emc2.core.instruments.KAZR objects. Among other radar and lidar instruments, EMC² includes instrument sub-classes for the full ARM radar and lidar instrument suite. Note that in the case of the KAZR class object (as in the case of different radar object) we specify the ARM site string to initialize the KAZR class object with the correct radar attributes.
# Set instrument class objects to simulate (a radar and a lidar)
KAZR = emc2.core.instruments.KAZR('nsa')
HSRL = emc2.core.instruments.HSRL()
print("Instrument class generation done!")
Instrument class generation done!
Initializing a Model Class Object#
Let’s take a look at the model output data file from EAM (E3SM’s atmospheric component) using the xarray Python package.
# Path to E3SM output file
model_path = './data/EMC2_demo_EAMv1_Freerun.2012-10-31.nc'
xr.open_dataset(model_path)
<xarray.Dataset>
Dimensions: (ncol_155w_to_158w_70n_to_73n: 3,
cosp_prs: 7, nbnd: 2,
cosp_tau: 7, cosp_scol: 10,
cosp_ht: 40, cosp_sr: 15,
cosp_sza: 5, cosp_htmisr: 16,
cosp_tau_modis: 6, lev: 72,
ilev: 73, time: 24)
Coordinates:
* cosp_prs (cosp_prs) float64 900.0 ... 90.0
* cosp_tau (cosp_tau) float64 0.15 ... 219.5
* cosp_scol (cosp_scol) int32 1 2 3 ... 8 9 10
* cosp_ht (cosp_ht) float64 240.0 ... 1.8...
* cosp_sr (cosp_sr) float64 0.605 ... 1.0...
* cosp_sza (cosp_sza) float64 0.0 ... 60.0
* cosp_htmisr (cosp_htmisr) float64 -99.0 ......
* cosp_tau_modis (cosp_tau_modis) float64 0.8 .....
* lev (lev) float64 0.1238 ... 998.5
* ilev (ilev) float64 0.1 ... 1e+03
* time (time) datetime64[ns] 2012-10-3...
Dimensions without coordinates: ncol_155w_to_158w_70n_to_73n, nbnd
Data variables: (12/206)
lat_155w_to_158w_70n_to_73n (ncol_155w_to_158w_70n_to_73n) float64 ...
lon_155w_to_158w_70n_to_73n (ncol_155w_to_158w_70n_to_73n) float64 ...
cosp_prs_bnds (cosp_prs, nbnd) float64 1e+03 ...
cosp_tau_bnds (cosp_tau, nbnd) float64 0.0 .....
cosp_ht_bnds (cosp_ht, nbnd) float64 0.0 ......
cosp_sr_bnds (cosp_sr, nbnd) float64 0.01 .....
... ...
soa_c2_155w_to_158w_70n_to_73n (time, lev, ncol_155w_to_158w_70n_to_73n) float32 ...
soa_c3_155w_to_158w_70n_to_73n (time, lev, ncol_155w_to_158w_70n_to_73n) float32 ...
wat_a1_155w_to_158w_70n_to_73n (time, lev, ncol_155w_to_158w_70n_to_73n) float32 ...
wat_a2_155w_to_158w_70n_to_73n (time, lev, ncol_155w_to_158w_70n_to_73n) float32 ...
wat_a3_155w_to_158w_70n_to_73n (time, lev, ncol_155w_to_158w_70n_to_73n) float32 ...
wat_a4_155w_to_158w_70n_to_73n (time, lev, ncol_155w_to_158w_70n_to_73n) float32 ...
Attributes:
Conventions: CF-1.0
source: CAM
case: EAMv1_ARM_Freerun_1
title: UNSET
logname: yshi
host: cori07
Version: $Name$
revision_Id: $Id$
initial_file: /global/cfs/cdirs/e3sm/inputdata/atm/cam/inic/homme/ca...
topography_file: /global/cfs/cdirs/e3sm/inputdata/atm/cam/topo/USGS-gto...
time_period_freq: hour_1- ncol_155w_to_158w_70n_to_73n: 3
- cosp_prs: 7
- nbnd: 2
- cosp_tau: 7
- cosp_scol: 10
- cosp_ht: 40
- cosp_sr: 15
- cosp_sza: 5
- cosp_htmisr: 16
- cosp_tau_modis: 6
- lev: 72
- ilev: 73
- time: 24
- cosp_prs(cosp_prs)float64900.0 740.0 620.0 ... 245.0 90.0
- long_name :
- COSP Mean ISCCP pressure
- units :
- hPa
- bounds :
- cosp_prs_bnds
array([900., 740., 620., 500., 375., 245., 90.])
- cosp_tau(cosp_tau)float640.15 0.8 2.45 6.5 16.2 41.5 219.5
- long_name :
- COSP Mean ISCCP optical depth
- units :
- 1
- bounds :
- cosp_tau_bnds
array([1.500e-01, 8.000e-01, 2.450e+00, 6.500e+00, 1.620e+01, 4.150e+01, 2.195e+02]) - cosp_scol(cosp_scol)int321 2 3 4 5 6 7 8 9 10
- long_name :
- COSP subcolumn
array([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10], dtype=int32)
- cosp_ht(cosp_ht)float64240.0 720.0 ... 1.848e+04 1.896e+04
- long_name :
- COSP Mean Height for lidar and radar simulator outputs
- units :
- m
- bounds :
- cosp_ht_bnds
array([ 240., 720., 1200., 1680., 2160., 2640., 3120., 3600., 4080., 4560., 5040., 5520., 6000., 6480., 6960., 7440., 7920., 8400., 8880., 9360., 9840., 10320., 10800., 11280., 11760., 12240., 12720., 13200., 13680., 14160., 14640., 15120., 15600., 16080., 16560., 17040., 17520., 18000., 18480., 18960.]) - cosp_sr(cosp_sr)float640.605 2.1 4.0 ... 539.5 1.004e+03
- long_name :
- COSP Mean Scattering Ratio for lidar simulator CFAD output
- units :
- 1
- bounds :
- cosp_sr_bnds
array([6.050e-01, 2.100e+00, 4.000e+00, 6.000e+00, 8.500e+00, 1.250e+01, 1.750e+01, 2.250e+01, 2.750e+01, 3.500e+01, 4.500e+01, 5.500e+01, 7.000e+01, 5.395e+02, 1.004e+03]) - cosp_sza(cosp_sza)float640.0 15.0 30.0 45.0 60.0
- long_name :
- COSP Parasol SZA
- units :
- degrees
array([ 0., 15., 30., 45., 60.])
- cosp_htmisr(cosp_htmisr)float64-99.0 0.25 0.75 ... 14.0 16.0 58.0
- long_name :
- COSP MISR height
- units :
- km
- bounds :
- cosp_htmisr_bnds
array([-99. , 0.25, 0.75, 1.25, 1.75, 2.25, 2.75, 3.5 , 4.5 , 6. , 8. , 10. , 12. , 14. , 16. , 58. ]) - cosp_tau_modis(cosp_tau_modis)float640.8 2.45 6.5 16.2 41.5 5.003e+04
- long_name :
- COSP Mean MODIS optical depth
- units :
- 1
- bounds :
- cosp_tau_modis_bnds
array([8.000e-01, 2.450e+00, 6.500e+00, 1.620e+01, 4.150e+01, 5.003e+04])
- lev(lev)float640.1238 0.1828 ... 993.8 998.5
- long_name :
- hybrid level at midpoints (1000*(A+B))
- units :
- hPa
- positive :
- down
- standard_name :
- atmosphere_hybrid_sigma_pressure_coordinate
- formula_terms :
- a: hyam b: hybm p0: P0 ps: PS
array([1.238254e-01, 1.828292e-01, 2.699489e-01, 3.985817e-01, 5.885091e-01, 8.689386e-01, 1.282995e+00, 1.894352e+00, 2.797027e+00, 4.129833e+00, 5.968449e+00, 8.377404e+00, 1.147379e+01, 1.533394e+01, 1.999634e+01, 2.544470e+01, 3.159325e+01, 3.836628e+01, 4.567120e+01, 5.330956e+01, 6.101518e+01, 6.847639e+01, 7.535534e+01, 8.194628e+01, 8.891054e+01, 9.646667e+01, 1.046650e+02, 1.135600e+02, 1.232110e+02, 1.336822e+02, 1.450433e+02, 1.573699e+02, 1.707441e+02, 1.852549e+02, 2.009989e+02, 2.180810e+02, 2.366148e+02, 2.567237e+02, 2.785416e+02, 3.022136e+02, 3.278975e+02, 3.557641e+02, 3.859990e+02, 4.188035e+02, 4.543958e+02, 4.924686e+02, 5.316395e+02, 5.706249e+02, 6.086438e+02, 6.453200e+02, 6.804980e+02, 7.137046e+02, 7.444748e+02, 7.723628e+02, 7.969527e+02, 8.178688e+02, 8.350952e+02, 8.496612e+02, 8.631764e+02, 8.763706e+02, 8.892227e+02, 9.017118e+02, 9.138175e+02, 9.255197e+02, 9.367990e+02, 9.476362e+02, 9.580128e+02, 9.679111e+02, 9.773141e+02, 9.862053e+02, 9.937570e+02, 9.984964e+02]) - ilev(ilev)float640.1 0.1477 0.218 ... 997.0 1e+03
- long_name :
- hybrid level at interfaces (1000*(A+B))
- units :
- hPa
- positive :
- down
- standard_name :
- atmosphere_hybrid_sigma_pressure_coordinate
- formula_terms :
- a: hyai b: hybi p0: P0 ps: PS
array([1.000000e-01, 1.476508e-01, 2.180076e-01, 3.218901e-01, 4.752733e-01, 7.017450e-01, 1.036132e+00, 1.529858e+00, 2.258847e+00, 3.335207e+00, 4.924460e+00, 7.012439e+00, 9.742370e+00, 1.320520e+01, 1.746267e+01, 2.253000e+01, 2.835939e+01, 3.482711e+01, 4.190545e+01, 4.943694e+01, 5.718218e+01, 6.484818e+01, 7.210460e+01, 7.860608e+01, 8.528648e+01, 9.253461e+01, 1.003987e+02, 1.089312e+02, 1.181888e+02, 1.282332e+02, 1.391312e+02, 1.509554e+02, 1.637844e+02, 1.777038e+02, 1.928061e+02, 2.091918e+02, 2.269702e+02, 2.462594e+02, 2.671880e+02, 2.898952e+02, 3.145321e+02, 3.412629e+02, 3.702654e+02, 4.017327e+02, 4.358743e+02, 4.729174e+02, 5.120198e+02, 5.512593e+02, 5.899905e+02, 6.272970e+02, 6.633429e+02, 6.976532e+02, 7.297561e+02, 7.591936e+02, 7.855321e+02, 8.083734e+02, 8.273643e+02, 8.428261e+02, 8.564964e+02, 8.698564e+02, 8.828849e+02, 8.955606e+02, 9.078631e+02, 9.197720e+02, 9.312675e+02, 9.423305e+02, 9.529419e+02, 9.630837e+02, 9.727385e+02, 9.818896e+02, 9.905210e+02, 9.969929e+02, 1.000000e+03]) - time(time)datetime64[ns]2012-10-31T01:00:00 ... 2012-11-01
- long_name :
- time
- bounds :
- time_bnds
array(['2012-10-31T01:00:00.000000000', '2012-10-31T02:00:00.000000000', '2012-10-31T03:00:00.000000000', '2012-10-31T04:00:00.000000000', '2012-10-31T05:00:00.000000000', '2012-10-31T06:00:00.000000000', '2012-10-31T07:00:00.000000000', '2012-10-31T08:00:00.000000000', '2012-10-31T09:00:00.000000000', '2012-10-31T10:00:00.000000000', '2012-10-31T11:00:00.000000000', '2012-10-31T12:00:00.000000000', '2012-10-31T13:00:00.000000000', '2012-10-31T14:00:00.000000000', '2012-10-31T15:00:00.000000000', '2012-10-31T16:00:00.000000000', '2012-10-31T17:00:00.000000000', '2012-10-31T18:00:00.000000000', '2012-10-31T19:00:00.000000000', '2012-10-31T20:00:00.000000000', '2012-10-31T21:00:00.000000000', '2012-10-31T22:00:00.000000000', '2012-10-31T23:00:00.000000000', '2012-11-01T00:00:00.000000000'], dtype='datetime64[ns]')
- lat_155w_to_158w_70n_to_73n(ncol_155w_to_158w_70n_to_73n)float64...
- long_name :
- latitude
- units :
- degrees_north
array([72.892828, 71.184778, 70.44341 ])
- lon_155w_to_158w_70n_to_73n(ncol_155w_to_158w_70n_to_73n)float64...
- long_name :
- longitude
- units :
- degrees_east
array([202.899162, 204.923226, 203.84069 ])
- cosp_prs_bnds(cosp_prs, nbnd)float64...
array([[1000., 800.], [ 800., 680.], [ 680., 560.], [ 560., 440.], [ 440., 310.], [ 310., 180.], [ 180., 0.]]) - cosp_tau_bnds(cosp_tau, nbnd)float64...
array([[0.00e+00, 3.00e-01], [3.00e-01, 1.30e+00], [1.30e+00, 3.60e+00], [3.60e+00, 9.40e+00], [9.40e+00, 2.30e+01], [2.30e+01, 6.00e+01], [6.00e+01, 3.79e+02]]) - cosp_ht_bnds(cosp_ht, nbnd)float64...
array([[ 0., 480.], [ 480., 960.], [ 960., 1440.], [ 1440., 1920.], [ 1920., 2400.], [ 2400., 2880.], [ 2880., 3360.], [ 3360., 3840.], [ 3840., 4320.], [ 4320., 4800.], [ 4800., 5280.], [ 5280., 5760.], [ 5760., 6240.], [ 6240., 6720.], [ 6720., 7200.], [ 7200., 7680.], [ 7680., 8160.], [ 8160., 8640.], [ 8640., 9120.], [ 9120., 9600.], [ 9600., 10080.], [10080., 10560.], [10560., 11040.], [11040., 11520.], [11520., 12000.], [12000., 12480.], [12480., 12960.], [12960., 13440.], [13440., 13920.], [13920., 14400.], [14400., 14880.], [14880., 15360.], [15360., 15840.], [15840., 16320.], [16320., 16800.], [16800., 17280.], [17280., 17760.], [17760., 18240.], [18240., 18720.], [18720., 19200.]]) - cosp_sr_bnds(cosp_sr, nbnd)float64...
array([[1.000e-02, 1.200e+00], [1.200e+00, 3.000e+00], [3.000e+00, 5.000e+00], [5.000e+00, 7.000e+00], [7.000e+00, 1.000e+01], [1.000e+01, 1.500e+01], [1.500e+01, 2.000e+01], [2.000e+01, 2.500e+01], [2.500e+01, 3.000e+01], [3.000e+01, 4.000e+01], [4.000e+01, 5.000e+01], [5.000e+01, 6.000e+01], [6.000e+01, 8.000e+01], [8.000e+01, 9.990e+02], [9.990e+02, 1.009e+03]]) - cosp_htmisr_bnds(cosp_htmisr, nbnd)float64...
array([[-99. , 0. ], [ 0. , 0.5], [ 0.5, 1. ], [ 1. , 1.5], [ 1.5, 2. ], [ 2. , 2.5], [ 2.5, 3. ], [ 3. , 4. ], [ 4. , 5. ], [ 5. , 7. ], [ 7. , 9. ], [ 9. , 11. ], [ 11. , 13. ], [ 13. , 15. ], [ 15. , 17. ], [ 17. , 99. ]]) - cosp_tau_modis_bnds(cosp_tau_modis, nbnd)float64...
array([[3.0e-01, 1.3e+00], [1.3e+00, 3.6e+00], [3.6e+00, 9.4e+00], [9.4e+00, 2.3e+01], [2.3e+01, 6.0e+01], [6.0e+01, 1.0e+05]]) - hyam(lev)float64...
- long_name :
- hybrid A coefficient at layer midpoints
array([1.238254e-04, 1.828292e-04, 2.699489e-04, 3.985817e-04, 5.885091e-04, 8.689386e-04, 1.282995e-03, 1.894352e-03, 2.797027e-03, 4.129833e-03, 5.968449e-03, 8.377404e-03, 1.147379e-02, 1.533394e-02, 1.999634e-02, 2.544470e-02, 3.159325e-02, 3.836628e-02, 4.567120e-02, 5.330956e-02, 6.101518e-02, 6.847639e-02, 7.535534e-02, 8.194628e-02, 8.891054e-02, 9.646667e-02, 1.046650e-01, 1.135600e-01, 1.232110e-01, 1.336822e-01, 1.450433e-01, 1.573699e-01, 1.707441e-01, 1.785448e-01, 1.775309e-01, 1.736633e-01, 1.694671e-01, 1.649142e-01, 1.599745e-01, 1.546149e-01, 1.487998e-01, 1.424905e-01, 1.356451e-01, 1.282178e-01, 1.201594e-01, 1.115393e-01, 1.026706e-01, 9.384396e-02, 8.523611e-02, 7.693226e-02, 6.896761e-02, 6.144932e-02, 5.448265e-02, 4.816853e-02, 4.260113e-02, 3.786552e-02, 3.396530e-02, 3.066740e-02, 2.760744e-02, 2.462015e-02, 2.171031e-02, 1.888266e-02, 1.614181e-02, 1.349230e-02, 1.093858e-02, 8.484941e-03, 6.135570e-03, 3.894491e-03, 1.765568e-03, 3.648085e-04, 0.000000e+00, 0.000000e+00]) - hybm(lev)float64...
- long_name :
- hybrid B coefficient at layer midpoints
array([0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0.00671 , 0.023468, 0.044418, 0.067148, 0.091809, 0.118567, 0.147599, 0.179098, 0.213274, 0.250354, 0.290586, 0.334236, 0.380929, 0.428969, 0.476781, 0.523408, 0.568388, 0.61153 , 0.652255, 0.689992, 0.724194, 0.754352, 0.780003, 0.80113 , 0.818994, 0.835569, 0.85175 , 0.867512, 0.882829, 0.897676, 0.912027, 0.92586 , 0.939151, 0.951877, 0.964017, 0.975548, 0.985841, 0.993757, 0.998496]) - P0()float64...
- long_name :
- reference pressure
- units :
- Pa
array(100000.)
- hyai(ilev)float64...
- long_name :
- hybrid A coefficient at layer interfaces
array([1.000000e-04, 1.476508e-04, 2.180076e-04, 3.218901e-04, 4.752733e-04, 7.017450e-04, 1.036132e-03, 1.529858e-03, 2.258847e-03, 3.335207e-03, 4.924460e-03, 7.012439e-03, 9.742370e-03, 1.320520e-02, 1.746267e-02, 2.253000e-02, 2.835939e-02, 3.482711e-02, 4.190545e-02, 4.943694e-02, 5.718218e-02, 6.484818e-02, 7.210460e-02, 7.860608e-02, 8.528648e-02, 9.253461e-02, 1.003987e-01, 1.089312e-01, 1.181888e-01, 1.282332e-01, 1.391312e-01, 1.509554e-01, 1.637844e-01, 1.777038e-01, 1.793858e-01, 1.756759e-01, 1.716507e-01, 1.672835e-01, 1.625450e-01, 1.574039e-01, 1.518259e-01, 1.457738e-01, 1.392073e-01, 1.320828e-01, 1.243528e-01, 1.159659e-01, 1.071127e-01, 9.822852e-02, 8.945939e-02, 8.101283e-02, 7.285170e-02, 6.508352e-02, 5.781512e-02, 5.115018e-02, 4.518688e-02, 4.001538e-02, 3.571566e-02, 3.221495e-02, 2.911986e-02, 2.609503e-02, 2.314527e-02, 2.027536e-02, 1.748996e-02, 1.479366e-02, 1.219095e-02, 9.686203e-03, 7.283674e-03, 4.987467e-03, 2.801519e-03, 7.296169e-04, 0.000000e+00, 0.000000e+00, 0.000000e+00]) - hybi(ilev)float64...
- long_name :
- hybrid B coefficient at layer interfaces
array([0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0.01342 , 0.033516, 0.055319, 0.078976, 0.104643, 0.132491, 0.162706, 0.195489, 0.231058, 0.26965 , 0.311521, 0.356951, 0.404907, 0.453031, 0.500531, 0.546284, 0.590491, 0.63257 , 0.671941, 0.708043, 0.740345, 0.768358, 0.791649, 0.810611, 0.827377, 0.843761, 0.85974 , 0.875285, 0.890373, 0.904978, 0.919077, 0.932644, 0.945658, 0.958096, 0.969937, 0.98116 , 0.990521, 0.996993, 1. ]) - date(time)int32...
- long_name :
- current date (YYYYMMDD)
array([20121031, 20121031, 20121031, 20121031, 20121031, 20121031, 20121031, 20121031, 20121031, 20121031, 20121031, 20121031, 20121031, 20121031, 20121031, 20121031, 20121031, 20121031, 20121031, 20121031, 20121031, 20121031, 20121031, 20121101], dtype=int32) - datesec(time)int32...
- long_name :
- current seconds of current date
array([ 3600, 7200, 10800, 14400, 18000, 21600, 25200, 28800, 32400, 36000, 39600, 43200, 46800, 50400, 54000, 57600, 61200, 64800, 68400, 72000, 75600, 79200, 82800, 0], dtype=int32) - time_bnds(time, nbnd)datetime64[ns]...
- long_name :
- time interval endpoints
array([['2012-10-31T00:00:00.000000000', '2012-10-31T01:00:00.000000000'], ['2012-10-31T01:00:00.000000000', '2012-10-31T02:00:00.000000000'], ['2012-10-31T02:00:00.000000000', '2012-10-31T03:00:00.000000000'], ['2012-10-31T03:00:00.000000000', '2012-10-31T04:00:00.000000000'], ['2012-10-31T04:00:00.000000000', '2012-10-31T05:00:00.000000000'], ['2012-10-31T05:00:00.000000000', '2012-10-31T06:00:00.000000000'], ['2012-10-31T06:00:00.000000000', '2012-10-31T07:00:00.000000000'], ['2012-10-31T07:00:00.000000000', '2012-10-31T08:00:00.000000000'], ['2012-10-31T08:00:00.000000000', '2012-10-31T09:00:00.000000000'], ['2012-10-31T09:00:00.000000000', '2012-10-31T10:00:00.000000000'], ['2012-10-31T10:00:00.000000000', '2012-10-31T11:00:00.000000000'], ['2012-10-31T11:00:00.000000000', '2012-10-31T12:00:00.000000000'], ['2012-10-31T12:00:00.000000000', '2012-10-31T13:00:00.000000000'], ['2012-10-31T13:00:00.000000000', '2012-10-31T14:00:00.000000000'], ['2012-10-31T14:00:00.000000000', '2012-10-31T15:00:00.000000000'], ['2012-10-31T15:00:00.000000000', '2012-10-31T16:00:00.000000000'], ['2012-10-31T16:00:00.000000000', '2012-10-31T17:00:00.000000000'], ['2012-10-31T17:00:00.000000000', '2012-10-31T18:00:00.000000000'], ['2012-10-31T18:00:00.000000000', '2012-10-31T19:00:00.000000000'], ['2012-10-31T19:00:00.000000000', '2012-10-31T20:00:00.000000000'], ['2012-10-31T20:00:00.000000000', '2012-10-31T21:00:00.000000000'], ['2012-10-31T21:00:00.000000000', '2012-10-31T22:00:00.000000000'], ['2012-10-31T22:00:00.000000000', '2012-10-31T23:00:00.000000000'], ['2012-10-31T23:00:00.000000000', '2012-11-01T00:00:00.000000000']], dtype='datetime64[ns]') - date_written(time)|S8...
array([b'01/12/22', b'01/12/22', b'01/12/22', b'01/12/22', b'01/12/22', b'01/12/22', b'01/12/22', b'01/12/22', b'01/12/22', b'01/12/22', b'01/12/22', b'01/12/22', b'01/12/22', b'01/12/22', b'01/12/22', b'01/12/22', b'01/12/22', b'01/12/22', b'01/12/22', b'01/12/22', b'01/12/22', b'01/12/22', b'01/12/22', b'01/12/22'], dtype='|S8') - time_written(time)|S8...
array([b'00:04:38', b'00:04:46', b'00:04:53', b'00:05:00', b'00:05:07', b'00:05:14', b'00:05:21', b'00:05:28', b'00:05:35', b'00:05:42', b'00:05:49', b'00:05:56', b'00:06:03', b'00:06:10', b'00:06:17', b'00:06:24', b'00:06:30', b'00:06:38', b'00:06:45', b'00:06:52', b'00:06:59', b'00:07:06', b'00:07:12', b'00:08:17'], dtype='|S8') - ndbase()int32...
- long_name :
- base day
array(0, dtype=int32)
- nsbase()int32...
- long_name :
- seconds of base day
array(0, dtype=int32)
- nbdate()int32...
- long_name :
- base date (YYYYMMDD)
array(20110101, dtype=int32)
- nbsec()int32...
- long_name :
- seconds of base date
array(0, dtype=int32)
- mdt()int32...
- long_name :
- timestep
- units :
- s
array(1800, dtype=int32)
- ndcur(time)int32...
- long_name :
- current day (from base day)
array([669, 669, 669, 669, 669, 669, 669, 669, 669, 669, 669, 669, 669, 669, 669, 669, 669, 669, 669, 669, 669, 669, 669, 670], dtype=int32) - nscur(time)int32...
- long_name :
- current seconds of current day
array([ 3600, 7200, 10800, 14400, 18000, 21600, 25200, 28800, 32400, 36000, 39600, 43200, 46800, 50400, 54000, 57600, 61200, 64800, 68400, 72000, 75600, 79200, 82800, 0], dtype=int32) - co2vmr(time)float64...
- long_name :
- co2 volume mixing ratio
array([0.000395, 0.000395, 0.000395, 0.000395, 0.000395, 0.000395, 0.000395, 0.000395, 0.000395, 0.000395, 0.000395, 0.000395, 0.000395, 0.000395, 0.000395, 0.000395, 0.000395, 0.000395, 0.000395, 0.000395, 0.000395, 0.000395, 0.000395, 0.000395]) - ch4vmr(time)float64...
- long_name :
- ch4 volume mixing ratio
array([1.821358e-06, 1.821359e-06, 1.821360e-06, 1.821361e-06, 1.821362e-06, 1.821363e-06, 1.821363e-06, 1.821364e-06, 1.821365e-06, 1.821366e-06, 1.821367e-06, 1.821368e-06, 1.821368e-06, 1.821369e-06, 1.821370e-06, 1.821371e-06, 1.821372e-06, 1.821373e-06, 1.821373e-06, 1.821374e-06, 1.821375e-06, 1.821376e-06, 1.821377e-06, 1.821378e-06]) - n2ovmr(time)float64...
- long_name :
- n2o volume mixing ratio
array([3.257664e-07, 3.257665e-07, 3.257666e-07, 3.257667e-07, 3.257668e-07, 3.257669e-07, 3.257670e-07, 3.257671e-07, 3.257672e-07, 3.257673e-07, 3.257674e-07, 3.257675e-07, 3.257676e-07, 3.257677e-07, 3.257678e-07, 3.257679e-07, 3.257680e-07, 3.257681e-07, 3.257682e-07, 3.257683e-07, 3.257684e-07, 3.257686e-07, 3.257687e-07, 3.257688e-07]) - f11vmr(time)float64...
- long_name :
- f11 volume mixing ratio
array([7.990421e-10, 7.990437e-10, 7.990452e-10, 7.990468e-10, 7.990483e-10, 7.990499e-10, 7.990514e-10, 7.990530e-10, 7.990545e-10, 7.990561e-10, 7.990576e-10, 7.990591e-10, 7.990607e-10, 7.990622e-10, 7.990638e-10, 7.990653e-10, 7.990669e-10, 7.990684e-10, 7.990700e-10, 7.990715e-10, 7.990731e-10, 7.990746e-10, 7.990761e-10, 7.990777e-10]) - f12vmr(time)float64...
- long_name :
- f12 volume mixing ratio
array([5.235691e-10, 5.235688e-10, 5.235685e-10, 5.235682e-10, 5.235678e-10, 5.235675e-10, 5.235672e-10, 5.235669e-10, 5.235666e-10, 5.235663e-10, 5.235660e-10, 5.235657e-10, 5.235654e-10, 5.235651e-10, 5.235647e-10, 5.235644e-10, 5.235641e-10, 5.235638e-10, 5.235635e-10, 5.235632e-10, 5.235629e-10, 5.235626e-10, 5.235623e-10, 5.235619e-10]) - sol_tsi(time)float64...
- long_name :
- total solar irradiance
- units :
- W/m2
array([-1., -1., -1., -1., -1., -1., -1., -1., -1., -1., -1., -1., -1., -1., -1., -1., -1., -1., -1., -1., -1., -1., -1., -1.]) - nsteph(time)int32...
- long_name :
- current timestep
array([32114, 32116, 32118, 32120, 32122, 32124, 32126, 32128, 32130, 32132, 32134, 32136, 32138, 32140, 32142, 32144, 32146, 32148, 32150, 32152, 32154, 32156, 32158, 32160], dtype=int32) - ACTNI_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- units :
- Micron
- long_name :
- Average Cloud Top ice number
- basename :
- ACTNI
array([[ 0. , 0. , 0. ], [ 8557.338 , 0. , 0. ], [ 2185.564 , 0. , 0. ], [ 3831.4524, 0. , 0. ], [ 3190.785 , 0. , 0. ], [ 8720.562 , 0. , 0. ], [ 20715.457 , 9476.448 , 3041.7554], [ 31962.346 , 6461.9336, 4046.7056], [ 34525.113 , 15306.996 , 9314.903 ], [ 28424.334 , 17412.85 , 9755.762 ], [ 18736.227 , 22751.783 , 13881.63 ], [ 10703.406 , 21750.72 , 18234.8 ], [ 3216.291 , 17995.281 , 16531.074 ], [ 1267.0157, 11974.873 , 12241.5625], [758922.8 , 5013.126 , 5331.2173], [ 19471.78 , 6522.024 , 1798.7648], [ 74358.58 , 16087.712 , 8368.193 ], [ 14553.429 , 14132.614 , 12102.414 ], [ 16908.633 , 8198.062 , 6487.5327], [ 18609.71 , 5317.317 , 3521.7761], [ 19512.51 , 4584.3867, 1490.0862], [ 3814.3347, 21297.027 , 20083.363 ], [ 41766.78 , 22866.082 , 21560.393 ], [ 44053.45 , 23709.127 , 22263.83 ]], dtype=float32) - ACTNL_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- units :
- Micron
- long_name :
- Average Cloud Top droplet number
- basename :
- ACTNL
array([[7.739711e+05, 7.676932e+05, 6.357267e+06], [0.000000e+00, 1.514196e+05, 1.121583e+03], [0.000000e+00, 3.925565e+06, 6.711506e+06], [0.000000e+00, 2.723435e+05, 7.043288e+06], [0.000000e+00, 2.437720e+06, 7.977614e+06], [0.000000e+00, 7.539384e+01, 3.300628e+06], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00]], dtype=float32) - ACTREI_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- units :
- Micron
- long_name :
- Average Cloud Top ice effective radius
- basename :
- ACTREI
array([[ 0. , 0. , 0. ], [29.185854, 0. , 0. ], [37.942253, 0. , 0. ], [40.049194, 0. , 0. ], [42.136127, 0. , 0. ], [40.377293, 0. , 0. ], [34.924427, 20.825615, 14.015091], [28.507236, 30.458511, 24.189386], [32.063515, 19.612564, 17.150776], [35.301277, 16.961077, 14.527979], [38.56587 , 20.120193, 16.700354], [40.704823, 29.697645, 25.212013], [42.44824 , 35.677147, 34.318676], [50.109352, 38.418274, 36.664425], [27.842411, 41.797993, 40.649006], [18.63569 , 43.067215, 43.85847 ], [37.700462, 39.503475, 41.8404 ], [50.358826, 35.682705, 37.789185], [49.45762 , 27.857752, 33.82564 ], [46.48117 , 18.007597, 24.24863 ], [40.129593, 46.73848 , 48.267353], [15.087814, 47.71531 , 48.236847], [50.0916 , 46.88475 , 47.694187], [71.42062 , 44.88017 , 46.677193]], dtype=float32) - ACTREL_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- units :
- Micron
- long_name :
- Average Cloud Top droplet effective radius
- basename :
- ACTREL
array([[ 9.533483, 20.934862, 1.100961], [ 0. , 28.352602, 1.456029], [ 0. , 7.656354, 17.115618], [ 0. , 6.00803 , 15.354406], [ 0. , 20.747929, 12.666198], [ 0. , 0.527036, 14.597877], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ]], dtype=float32) - ADRAIN_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- Micron
- long_name :
- Average rain effective Diameter
- basename :
- ADRAIN
array([[[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [8.528968e-05, 2.000000e-05, 2.689993e-05], [1.114486e-04, 2.793106e-05, 2.572727e-05]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [8.372214e-05, 2.153700e-05, 2.913216e-05], [1.097475e-04, 0.000000e+00, 2.778940e-05]], ..., [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [8.794744e-05, 8.332219e-05, 0.000000e+00], [1.048377e-04, 9.801649e-05, 0.000000e+00]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [8.291411e-05, 8.704309e-05, 9.624913e-05], [1.001206e-04, 1.016431e-04, 1.138372e-04]]], dtype=float32) - ADSNOW_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- Micron
- long_name :
- Average snow effective Diameter
- basename :
- ADSNOW
array([[[0. , 0. , 0. ], [0. , 0. , 0. ], ..., [0. , 0.000374, 0.000262], [0. , 0.000376, 0.000288]], [[0. , 0. , 0. ], [0. , 0. , 0. ], ..., [0. , 0.00038 , 0.000267], [0. , 0.00038 , 0.000293]], ..., [[0. , 0. , 0. ], [0. , 0. , 0. ], ..., [0.000804, 0.000724, 0.002056], [0.000857, 0.00076 , 0.002446]], [[0. , 0. , 0. ], [0. , 0. , 0. ], ..., [0.000765, 0.000675, 0.000806], [0.000809, 0.000708, 0.000875]]], dtype=float32) - ANRAIN_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- m-3
- long_name :
- Average rain number conc
- basename :
- ANRAIN
array([[[ 0. , 0. , 0. ], [ 0. , 0. , 0. ], ..., [ 1.030104, 0.842169, 33.427464], [ 0.701162, 0.656185, 2.889647]], [[ 0. , 0. , 0. ], [ 0. , 0. , 0. ], ..., [ 0.850479, 1.066058, 42.5439 ], [ 0.577835, 0.582804, 3.437178]], ..., [[ 0. , 0. , 0. ], [ 0. , 0. , 0. ], ..., [ 1.488433, 2.963411, 0. ], [ 0.815862, 0.976173, 0. ]], [[ 0. , 0. , 0. ], [ 0. , 0. , 0. ], ..., [ 1.600822, 1.990333, 0.138441], [ 0.843817, 0.650359, 0.086979]]], dtype=float32) - ANSNOW_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- m-3
- long_name :
- Average snow number conc
- basename :
- ANSNOW
array([[[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [0.000000e+00, 5.187287e-03, 1.469719e+00], [0.000000e+00, 4.119792e-03, 1.215414e-01]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [0.000000e+00, 5.532943e-03, 1.933517e+00], [0.000000e+00, 4.358483e-03, 1.422343e-01]], ..., [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [3.277525e-02, 1.938923e-01, 1.563086e-03], [2.396860e-02, 8.391221e-02, 1.118287e-03]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [2.725423e-02, 1.315075e-01, 1.890629e-02], [2.040495e-02, 5.602907e-02, 1.632102e-02]]], dtype=float32) - AODVIS_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- long_name :
- Aerosol optical depth 550 nm
- basename :
- AODVIS
array([[0.030712, 0.032778, 0.033396], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, 0.028919], [0.114285, 0.044971, 0.033484], [0.097061, 0.0633 , 0.040738], [0.091285, 0.064523, 0.051156], [0.109229, 0.092784, 0.070763], [0.128933, 0.090308, 0.087872]], dtype=float32) - AQRAIN_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Average rain mixing ratio
- basename :
- AQRAIN
array([[[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [1.914412e-10, 4.179978e-11, 1.950170e-09], [1.479660e-10, 2.502543e-11, 1.469315e-10]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [1.473782e-10, 4.627196e-11, 3.146754e-09], [1.140054e-10, 2.260730e-11, 2.278695e-10]], ..., [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [1.229262e-09, 2.135960e-09, 0.000000e+00], [9.373496e-10, 9.364270e-10, 0.000000e+00]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [1.233039e-09, 1.708460e-09, 1.989750e-10], [9.327485e-10, 7.383295e-10, 1.778090e-10]]], dtype=float32) - AQSNOW_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Average snow mixing ratio
- basename :
- AQSNOW
array([[[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [0.000000e+00, 1.436742e-10, 1.615973e-08], [0.000000e+00, 1.251544e-10, 1.558078e-09]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [0.000000e+00, 1.575783e-10, 2.271958e-08], [0.000000e+00, 1.352032e-10, 1.964119e-09]], ..., [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [8.182279e-09, 4.193304e-08, 5.477341e-09], [6.654794e-09, 1.957000e-08, 5.465002e-09]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [5.982915e-09, 2.167550e-08, 5.790207e-09], [4.889019e-09, 9.927276e-09, 5.788096e-09]]], dtype=float32) - AREI_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- Micron
- long_name :
- Average ice effective radius
- basename :
- AREI
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - AREL_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- Micron
- long_name :
- Average droplet effective radius
- basename :
- AREL
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - AWNC_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- m-3
- long_name :
- Average cloud water number conc
- basename :
- AWNC
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - AWNI_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- m-3
- long_name :
- Average cloud ice number conc
- basename :
- AWNI
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - CCN3_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- #/cm3
- long_name :
- CCN concentration at S=0.1%
- basename :
- CCN3
array([[[ 0. , 0. , 0. ], [ 0. , 0. , 0. ], ..., [10.027431, 9.827665, 10.880567], [10.087448, 9.887123, 10.952791]], [[ 0. , 0. , 0. ], [ 0. , 0. , 0. ], ..., [ 9.983613, 9.482944, 10.61025 ], [10.045017, 9.539835, 10.676319]], ..., [[ 0. , 0. , 0. ], [ 0. , 0. , 0. ], ..., [18.529238, 18.205244, 13.857922], [18.690313, 18.332838, 13.983971]], [[ 0. , 0. , 0. ], [ 0. , 0. , 0. ], ..., [17.542122, 18.641277, 12.998683], [17.697273, 18.781399, 13.140232]]], dtype=float32) - CDNUMC_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- units :
- 1/m2
- long_name :
- Vertically-integrated droplet concentration
- basename :
- CDNUMC
array([[1.168949e+10, 1.565143e+10, 1.601807e+10], [1.185835e+10, 1.411084e+10, 1.400212e+10], [1.222992e+10, 1.444714e+10, 1.348254e+10], [1.300572e+10, 1.327612e+10, 1.333480e+10], [1.322750e+10, 1.277352e+10, 1.303767e+10], [1.365469e+10, 1.341201e+10, 1.133901e+10], [1.510588e+10, 1.401146e+10, 1.202419e+10], [1.607141e+10, 1.433679e+10, 1.179170e+10], [1.438302e+10, 1.353384e+10, 9.897816e+09], [1.716804e+10, 1.205266e+10, 9.658508e+09], [3.114484e+10, 9.949229e+09, 9.211769e+09], [2.938556e+10, 7.209252e+09, 4.744780e+09], [4.367438e+10, 6.079790e+09, 1.233411e-03], [4.232584e+10, 5.940761e+09, 1.045398e-08], [4.469054e+10, 3.666620e+09, 1.045029e-08], [4.585252e+10, 1.138172e+10, 1.044508e-08], [3.973812e+10, 2.574661e+10, 4.339235e+09], [1.790189e+10, 2.433514e+10, 1.773920e+10], [1.348755e+10, 1.868215e+10, 1.274098e+10], [1.446943e+10, 1.507829e+10, 1.203071e+10], [3.344169e+10, 1.993745e+10, 1.271975e+10], [3.438628e+10, 3.333803e+10, 2.196158e+10], [2.880439e+10, 3.074996e+10, 3.624091e+10], [3.066245e+10, 2.284805e+10, 2.560258e+10]], dtype=float32) - CLDHGH_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- units :
- fraction
- long_name :
- Vertically-integrated high cloud
- basename :
- CLDHGH
array([[0.00104 , 0. , 0. ], [0.808379, 0. , 0. ], [0.999 , 0. , 0. ], [0.999 , 0.003357, 0. ], [0.999 , 0.143784, 0.048882], [0.999 , 0.410026, 0.195918], [0.999 , 0.890796, 0.508126], [0.999 , 0.889183, 0.751768], [0.999 , 0.693581, 0.625902], [0.999 , 0.632163, 0.560871], [0.999 , 0.749432, 0.631077], [0.999 , 0.917096, 0.82272 ], [0.999 , 0.999 , 0.998233], [0.999 , 0.999 , 0.999 ], [0.999 , 0.999 , 0.999 ], [0.999 , 0.999 , 0.999 ], [0.999 , 0.999 , 0.999 ], [0.999 , 0.999 , 0.999 ], [0.999 , 0.999 , 0.999 ], [0.999 , 0.999 , 0.999 ], [0.999 , 0.999 , 0.999 ], [0.809342, 0.999 , 0.999 ], [0.078824, 0.999 , 0.999 ], [0. , 0.999 , 0.999 ]], dtype=float32) - CLDICE_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Grid box averaged cloud ice amount
- basename :
- CLDICE
array([[[3.696545e-24, 3.664190e-24, 3.697063e-24], [3.861715e-24, 3.707140e-24, 3.702022e-24], ..., [1.902475e-13, 8.940100e-15, 6.967840e-13], [4.922656e-15, 1.406920e-14, 3.357862e-16]], [[3.751631e-24, 3.676623e-24, 3.692936e-24], [3.865746e-24, 3.746652e-24, 3.754342e-24], ..., [1.551619e-14, 7.817670e-15, 2.259932e-12], [2.948848e-16, 1.280838e-14, 4.156469e-16]], ..., [[3.760485e-24, 3.728816e-24, 3.713117e-24], [3.983544e-24, 3.975421e-24, 3.937411e-24], ..., [1.180629e-16, 6.376558e-16, 2.405415e-14], [9.963031e-17, 3.006204e-16, 2.689951e-14]], [[3.822396e-24, 3.778702e-24, 3.773467e-24], [4.002707e-24, 3.956122e-24, 3.885373e-24], ..., [1.142677e-16, 7.229459e-16, 3.754779e-14], [9.820425e-17, 1.111049e-16, 5.539376e-14]]], dtype=float32) - CLDLIQ_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Grid box averaged cloud liquid amount
- basename :
- CLDLIQ
array([[[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [1.990015e-35, 0.000000e+00, 0.000000e+00], [4.305099e-36, 0.000000e+00, 0.000000e+00]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [8.887747e-38, 0.000000e+00, 0.000000e+00], [8.215617e-38, 0.000000e+00, 0.000000e+00]], ..., [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00]]], dtype=float32) - CLDLOW_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- units :
- fraction
- long_name :
- Vertically-integrated low cloud
- basename :
- CLDLOW
array([[1.000000e+00, 9.999881e-01, 9.999075e-01], [9.999999e-01, 9.999717e-01, 9.998953e-01], [1.000000e+00, 9.999469e-01, 9.999406e-01], [1.000000e+00, 9.999158e-01, 1.000000e+00], [1.000000e+00, 9.998755e-01, 9.999391e-01], [1.000000e+00, 9.999039e-01, 9.999171e-01], [9.999999e-01, 9.999537e-01, 9.999434e-01], [1.000000e+00, 9.999146e-01, 9.997387e-01], [9.999927e-01, 9.998155e-01, 9.997863e-01], [9.999965e-01, 9.996963e-01, 1.000000e+00], [1.000000e+00, 9.990000e-01, 1.000000e+00], [9.999933e-01, 9.990000e-01, 1.000000e+00], [1.000000e+00, 1.000000e+00, 9.990000e-01], [9.999906e-01, 1.000000e+00, 9.990000e-01], [1.000000e+00, 6.992201e-01, 4.926284e-02], [1.000000e+00, 1.000000e+00, 0.000000e+00], [1.000000e+00, 1.000000e+00, 0.000000e+00], [1.000000e+00, 1.000000e+00, 1.339780e-05], [1.000000e+00, 8.989547e-01, 5.853169e-01], [1.000000e+00, 9.994234e-01, 9.990000e-01], [1.000000e+00, 1.000000e+00, 1.000000e+00], [1.000000e+00, 1.000000e+00, 1.000000e+00], [1.000000e+00, 1.000000e+00, 1.000000e+00], [1.000000e+00, 1.000000e+00, 1.000000e+00]], dtype=float32) - CLDMED_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- units :
- fraction
- long_name :
- Vertically-integrated mid-level cloud
- basename :
- CLDMED
array([[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [1.335267e-01, 0.000000e+00, 0.000000e+00], [5.794461e-01, 0.000000e+00, 0.000000e+00], [9.990000e-01, 0.000000e+00, 0.000000e+00], [9.990000e-01, 0.000000e+00, 0.000000e+00], [9.990000e-01, 0.000000e+00, 0.000000e+00], [9.990000e-01, 8.403970e-02, 5.026031e-05], [9.990000e-01, 5.139107e-01, 1.308315e-01], [1.000000e+00, 9.990000e-01, 5.585143e-01], [1.000000e+00, 9.990000e-01, 9.990000e-01], [1.000000e+00, 9.990000e-01, 9.990000e-01], [1.000000e+00, 9.990000e-01, 9.990000e-01], [1.000000e+00, 9.990000e-01, 9.990000e-01], [1.000000e+00, 1.000000e+00, 9.990000e-01], [1.000000e+00, 1.000000e+00, 9.990000e-01], [1.000000e+00, 1.000000e+00, 1.000000e+00], [1.000000e+00, 1.000000e+00, 9.999743e-01], [1.000000e+00, 1.000000e+00, 1.000000e+00], [1.000000e+00, 1.000000e+00, 1.000000e+00], [1.000000e+00, 1.000000e+00, 1.000000e+00], [1.000000e+00, 1.000000e+00, 1.000000e+00], [1.000000e+00, 1.000000e+00, 1.000000e+00]], dtype=float32) - CLDTOT_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- units :
- fraction
- long_name :
- Vertically-integrated total cloud
- basename :
- CLDTOT
array([[1. , 0.999988, 0.999907], [1. , 0.999972, 0.999895], [1. , 0.999947, 0.999941], [1. , 0.999916, 1. ], [1. , 0.999875, 0.999939], [1. , 0.999904, 0.999917], [1. , 0.999954, 0.999943], [1. , 0.999915, 0.999739], [0.999993, 0.999815, 0.999786], [0.999997, 0.999696, 1. ], [1. , 0.999 , 1. ], [1. , 0.999 , 1. ], [1. , 1. , 0.999 ], [1. , 1. , 0.999 ], [1. , 0.999 , 0.999 ], [1. , 1. , 0.999 ], [1. , 1. , 0.999 ], [1. , 1. , 1. ], [1. , 1. , 0.999974], [1. , 1. , 1. ], [1. , 1. , 1. ], [1. , 1. , 1. ], [1. , 1. , 1. ], [1. , 1. , 1. ]], dtype=float32) - CLOUD_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- fraction
- long_name :
- Cloud fraction
- basename :
- CLOUD
array([[[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [6.295952e-05, 1.000000e-04, 3.829989e-03], [2.373634e-05, 1.000000e-04, 5.159241e-04]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [6.425572e-05, 1.000000e-04, 5.184209e-03], [2.275802e-05, 1.000000e-04, 6.358125e-04]], ..., [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [1.231354e-04, 6.401844e-04, 0.000000e+00], [5.586237e-05, 3.004619e-04, 0.000000e+00]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [1.221820e-04, 2.557086e-04, 0.000000e+00], [5.453656e-05, 1.176522e-04, 0.000000e+00]]], dtype=float32) - CMFDICE_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg/s
- long_name :
- Cloud ice tendency - shallow convection
- basename :
- CMFDICE
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - CMFDLIQ_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg/s
- long_name :
- Cloud liq tendency - shallow convection
- basename :
- CMFDLIQ
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - CMFDQ_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg/s
- long_name :
- QV tendency - shallow convection
- basename :
- CMFDQ
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - CMFDT_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- K/s
- long_name :
- T tendency - shallow convection
- basename :
- CMFDT
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - CONCLD_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- fraction
- long_name :
- Convective cloud cover
- basename :
- CONCLD
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - EMIS_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- 1
- long_name :
- Cloud longwave emissivity
- basename :
- EMIS
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - EVAPPREC_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg/s
- long_name :
- Rate of evaporation of falling precip
- basename :
- EVAPPREC
array([[[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [6.370819e-09, 0.000000e+00, 6.069387e-09], [4.923416e-09, 0.000000e+00, 3.932127e-09]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [4.905917e-09, 0.000000e+00, 6.099817e-09], [3.794396e-09, 0.000000e+00, 3.843893e-09]], ..., [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [8.506556e-09, 3.645129e-09, 1.478083e-10], [7.374191e-09, 3.155973e-09, 1.850277e-10]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [9.433403e-09, 3.561139e-09, 5.810529e-10], [7.662142e-09, 3.429365e-09, 7.995131e-10]]], dtype=float32) - EVAPSNOW_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg/s
- long_name :
- Rate of evaporation of falling snow
- basename :
- EVAPSNOW
array([[[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [3.831174e-13, 0.000000e+00, 4.578196e-09], [4.803308e-13, 0.000000e+00, 3.322691e-09]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [3.131284e-13, 0.000000e+00, 4.327630e-09], [3.914658e-13, 0.000000e+00, 3.102591e-09]], ..., [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [2.770460e-09, 3.311239e-09, 1.454018e-10], [2.832158e-09, 2.986141e-09, 1.831395e-10]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [2.435393e-09, 2.919134e-09, 5.237805e-10], [2.528656e-09, 2.662716e-09, 7.616963e-10]]], dtype=float32) - FCTI_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- units :
- fraction
- long_name :
- Fractional occurrence of cloud top ice
- basename :
- FCTI
array([[0. , 0. , 0. ], [0.808379, 0. , 0. ], [0.999 , 0. , 0. ], [0.999 , 0. , 0. ], [0.999 , 0. , 0. ], [0.999 , 0. , 0. ], [0.999 , 0.678768, 0.508126], [0.895923, 0.889183, 0.751768], [0.977181, 0.693581, 0.625902], [0.999 , 0.632163, 0.560871], [0.999 , 0.721187, 0.626666], [0.999 , 0.917096, 0.82272 ], [0.999 , 0.999 , 0.998233], [0.999 , 0.999 , 0.999 ], [0.999 , 0.999 , 0.999 ], [0.66232 , 0.999 , 0.999 ], [0.999 , 0.999 , 0.999 ], [0.999 , 0.999 , 0.999 ], [0.999 , 0.84591 , 0.963859], [0.999 , 0.661171, 0.776357], [0.997783, 0.999 , 0.999 ], [0.52857 , 0.999 , 0.999 ], [0.999 , 0.999 , 0.999 ], [0.999 , 0.999 , 0.999 ]], dtype=float32) - FCTL_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- units :
- fraction
- long_name :
- Fractional occurrence of cloud top liquid
- basename :
- FCTL
array([[0.431687, 0.999 , 0.189045], [0. , 0.999 , 0.050763], [0. , 0.999 , 0.999 ], [0. , 0.771017, 1. ], [0. , 0.999 , 0.999 ], [0. , 0.018375, 0.999 ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ]], dtype=float32) - FDL_155w_to_158w_70n_to_73n(time, ilev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 10
- units :
- W/m2
- long_name :
- Longwave downward flux
- basename :
- FDL
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - FDLC_155w_to_158w_70n_to_73n(time, ilev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 10
- units :
- W/m2
- long_name :
- Longwave clear-sky downward flux
- basename :
- FDLC
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - FDS_155w_to_158w_70n_to_73n(time, ilev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 10
- units :
- W/m2
- long_name :
- Shortwave downward flux
- basename :
- FDS
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - FDSC_155w_to_158w_70n_to_73n(time, ilev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 10
- units :
- W/m2
- long_name :
- Shortwave clear-sky downward flux
- basename :
- FDSC
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - FICE_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- fraction
- long_name :
- Fractional ice content within cloud
- basename :
- FICE
array([[[0. , 0. , 0. ], [0. , 0. , 0. ], ..., [0.001034, 0.774633, 0.892315], [0.001445, 0.833364, 0.913824]], [[0. , 0. , 0. ], [0. , 0. , 0. ], ..., [0.001115, 0.77301 , 0.878346], [0.001594, 0.856744, 0.896044]], ..., [[0. , 0. , 0. ], [0. , 0. , 0. ], ..., [0.869388, 0.951531, 0.99823 ], [0.876537, 0.954335, 0.998496]], [[0. , 0. , 0. ], [0. , 0. , 0. ], ..., [0.829123, 0.926939, 0.966778], [0.839783, 0.930775, 0.970196]]], dtype=float32) - FLDS_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Downwelling longwave flux at surface
- basename :
- FLDS
array([[280.69238, 279.5848 , 268.81247], [280.73898, 280.55875, 269.60934], [280.73428, 281.1117 , 270.55765], [280.62527, 281.5819 , 270.97412], [280.52295, 282.0952 , 271.36642], [280.24716, 282.04788, 271.561 ], [280.0014 , 281.7211 , 271.31067], [279.72656, 281.2767 , 270.81897], [279.54318, 280.09525, 269.73972], [279.8138 , 278.20303, 268.45862], [280.28513, 276.70023, 264.67325], [281.11362, 272.76822, 242.96251], [282.88794, 259.34152, 225.07799], [285.0478 , 260.79016, 213.4504 ], [288.19315, 256.55664, 219.29738], [291.51593, 251.37383, 223.65433], [294.03168, 262.5557 , 233.09926], [296.73447, 261.82407, 249.48769], [299.87808, 270.3087 , 256.61465], [300.401 , 274.92416, 264.2902 ], [301.54843, 280.39655, 264.72757], [301.96033, 287.54373, 270.38773], [302.12268, 291.24042, 278.07443], [301.83682, 294.20407, 283.38895]], dtype=float32) - FLDSC_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Clearsky Downwelling longwave flux at surface
- basename :
- FLDSC
array([[199.45505, 200.6656 , 194.24307], [199.69139, 201.43248, 194.76727], [199.83374, 202.09734, 195.29198], [200.00172, 202.71983, 195.91995], [200.0962 , 203.1552 , 196.39651], [200.31114, 203.44568, 196.76009], [200.65015, 203.58481, 196.96777], [201.28447, 203.5882 , 196.96017], [202.20958, 203.50243, 196.79395], [203.43958, 203.33086, 196.4611 ], [205.04779, 203.0933 , 196.06673], [207.01964, 202.8593 , 195.42273], [209.29767, 202.74529, 193.59747], [211.78474, 202.68143, 191.9114 ], [214.36734, 203.05722, 190.87985], [216.90399, 203.69518, 191.37105], [219.42882, 204.6364 , 192.31377], [221.62253, 205.93706, 193.70764], [223.50446, 207.32422, 195.67503], [225.09474, 209.00395, 197.58386], [226.09483, 210.97305, 199.87434], [226.7545 , 213.0335 , 202.43811], [227.0952 , 215.491 , 205.46333], [226.95184, 217.80408, 208.5933 ]], dtype=float32) - FLN200_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Net longwave flux at 200 mb
- basename :
- FLN200
array([[181.63145, 175.78432, 171.92537], [181.46632, 176.63123, 172.33615], [180.69075, 177.01093, 172.84773], [179.61081, 177.35898, 173.32806], [178.48746, 177.6213 , 173.68597], [173.12837, 177.5207 , 174.12689], [165.3101 , 177.09372, 174.35172], [160.04568, 176.44109, 174.25577], [156.30577, 175.96399, 173.99644], [154.451 , 174.70705, 173.63492], [149.07567, 171.8142 , 173.09581], [144.46935, 167.69159, 174.99707], [142.95679, 165.44112, 170.55946], [140.55365, 161.28178, 165.46378], [140.0747 , 160.37535, 161.35063], [130.60341, 161.53316, 161.7308 ], [134.4604 , 142.10281, 150.46484], [133.2863 , 136.21169, 136.69405], [140.45877, 134.67688, 138.03511], [145.63287, 139.2056 , 142.445 ], [141.6708 , 142.80418, 144.25475], [135.6937 , 146.34378, 146.19223], [139.82283, 138.442 , 140.51271], [147.79951, 137.21866, 137.94554]], dtype=float32) - FLNS_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Net longwave flux at surface
- basename :
- FLNS
array([[33.262386, 28.076458, 14.879767], [33.19853 , 27.268536, 13.857829], [33.18599 , 26.927505, 13.060074], [33.27775 , 26.659029, 13.101119], [33.362812, 26.308744, 13.020462], [33.62132 , 26.465555, 12.980641], [33.849827, 26.815142, 13.171859], [34.10743 , 27.19768 , 13.214188], [34.273537, 28.23514 , 13.407933], [33.985653, 29.84681 , 13.408607], [33.497086, 30.969513, 15.774937], [32.651333, 34.653973, 35.297207], [30.859768, 47.881855, 43.409813], [28.682705, 45.608395, 46.090366], [25.520098, 50.369427, 35.262463], [22.180082, 55.572086, 35.103825], [19.647112, 44.417088, 30.257277], [16.927076, 45.848354, 18.36789 ], [13.766231, 37.49404 , 16.225924], [13.226058, 33.354782, 11.236076], [12.061398, 28.291662, 13.382421], [11.632226, 21.743204, 9.650012], [11.452618, 18.738386, 4.758803], [11.721215, 16.44939 , 2.738184]], dtype=float32) - FLNT_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Net longwave flux at top of model
- basename :
- FLNT
array([[200.90723, 195.60602, 192.05638], [200.71204, 196.39726, 192.42897], [199.9027 , 196.71967, 192.8777 ], [198.78297, 197.01344, 193.29637], [197.6929 , 197.25195, 193.62509], [192.50615, 197.1396 , 194.0176 ], [185.00961, 196.67488, 194.18846], [179.83078, 195.96284, 194.0043 ], [176.1282 , 195.41545, 193.65614], [174.2608 , 194.1051 , 193.21149], [168.96216, 191.24405, 192.58568], [164.48337, 187.25597, 194.25847], [162.92906, 184.89346, 189.98956], [160.5387 , 180.83022, 185.10435], [159.99771, 179.94891, 181.02621], [150.87563, 180.9767 , 181.29364], [154.43205, 162.226 , 170.38597], [153.26256, 156.63591, 157.17462], [160.05908, 155.03209, 158.38156], [164.94113, 159.28558, 162.58072], [161.00629, 162.63834, 164.23163], [155.25272, 165.94565, 165.99251], [159.16544, 158.2671 , 160.47804], [166.81964, 157.05074, 157.96744]], dtype=float32) - FLUT_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Upwelling longwave flux at top of model
- basename :
- FLUT
array([[201.19269, 195.91656, 192.37566], [201.00763, 196.72093, 192.76154], [200.19627, 197.04715, 193.21114], [199.06737, 197.33434, 193.62299], [197.96565, 197.5612 , 193.94482], [192.76982, 197.43474, 194.3252 ], [185.26625, 196.96098, 194.48659], [180.08305, 196.24464, 194.29663], [176.3774 , 195.69421, 193.94502], [174.50429, 194.3774 , 193.49557], [169.19966, 191.50334, 192.8557 ], [164.71968, 187.50366, 194.51598], [163.166 , 185.13377, 190.23761], [160.7791 , 181.06902, 185.34995], [160.24316, 180.19089, 181.27469], [151.12617, 181.2226 , 181.54636], [154.68646, 162.4738 , 170.63956], [153.5171 , 156.88481, 157.42708], [160.31122, 155.28313, 158.63702], [165.19371, 159.54097, 162.84389], [161.2642 , 162.90495, 164.50737], [155.51704, 166.22649, 166.27759], [159.434 , 158.55186, 160.76585], [167.08875, 157.33684, 158.25694]], dtype=float32) - FLUTC_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Clearsky upwelling longwave flux at top of model
- basename :
- FLUTC
array([[222.91275, 222.00468, 213.85579], [222.63602, 221.90807, 213.67801], [222.31342, 221.77852, 213.57503], [221.95903, 221.64247, 213.54716], [221.55283, 221.48502, 213.44911], [221.12857, 221.24738, 213.23395], [220.76405, 220.92285, 212.88655], [220.37428, 220.55666, 212.36052], [220.02591, 220.14854, 211.68695], [219.67354, 219.66013, 210.86441], [219.31143, 219.10959, 209.93048], [219.05827, 218.59978, 208.71603], [218.86418, 218.17699, 204.88757], [218.67836, 217.58351, 201.41728], [218.61922, 217.45003, 199.41467], [218.81314, 217.10815, 200.6058 ], [219.00188, 216.90114, 202.06958], [219.58716, 217.05151, 203.64798], [220.07086, 217.46791, 205.44212], [220.14432, 217.79782, 206.75606], [220.10417, 217.91089, 207.8027 ], [220.27509, 218.02267, 208.39905], [220.46239, 218.16553, 209.34784], [220.71407, 218.57372, 210.57094]], dtype=float32) - FREQI_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- fraction
- long_name :
- Fractional occurrence of ice
- basename :
- FREQI
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - FREQL_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- fraction
- long_name :
- Fractional occurrence of liquid
- basename :
- FREQL
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - FREQR_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- fraction
- long_name :
- Fractional occurrence of rain
- basename :
- FREQR
array([[[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [1.000000e-04, 1.000000e-04, 3.829989e-03], [1.000000e-04, 1.000000e-04, 5.159241e-04]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [1.000000e-04, 1.000000e-04, 5.184209e-03], [1.000000e-04, 1.000000e-04, 6.358125e-04]], ..., [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [1.231354e-04, 6.401844e-04, 0.000000e+00], [1.000000e-04, 3.004619e-04, 0.000000e+00]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [1.221820e-04, 2.557086e-04, 1.000000e-04], [1.000000e-04, 1.176522e-04, 1.000000e-04]]], dtype=float32) - FREQS_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- fraction
- long_name :
- Fractional occurrence of snow
- basename :
- FREQS
array([[[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [0.000000e+00, 1.000000e-04, 3.829989e-03], [0.000000e+00, 1.000000e-04, 5.159241e-04]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [0.000000e+00, 1.000000e-04, 5.184209e-03], [0.000000e+00, 1.000000e-04, 6.358125e-04]], ..., [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [1.231354e-04, 6.401844e-04, 1.000000e-04], [1.000000e-04, 3.004619e-04, 1.000000e-04]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [1.221820e-04, 2.557086e-04, 1.000000e-04], [1.000000e-04, 1.176522e-04, 1.000000e-04]]], dtype=float32) - FSDS_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Downwelling solar flux at surface
- basename :
- FSDS
array([[2.678406e-01, 1.545140e+00, 5.795152e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 1.611715e-03], [4.063277e-01, 7.903570e+00, 8.939452e+00], [3.172492e+00, 1.415591e+01, 2.568836e+01], [5.056305e+00, 1.624425e+01, 3.229261e+01], [4.639563e+00, 1.257831e+01, 2.841950e+01], [2.174334e+00, 7.520370e+00, 1.671395e+01]], dtype=float32) - FSDSC_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Clearsky downwelling solar flux at surface
- basename :
- FSDSC
array([[7.079148e-01, 4.409168e+00, 1.067069e+01], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 1.803833e-03], [5.036520e-01, 9.187177e+00, 1.350483e+01], [1.099571e+01, 3.397618e+01, 4.469991e+01], [2.256600e+01, 4.840746e+01, 6.196995e+01], [2.174504e+01, 4.403760e+01, 5.864518e+01], [9.655072e+00, 2.490135e+01, 3.710740e+01]], dtype=float32) - FSNS_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Net solar flux at surface
- basename :
- FSNS
array([[ 0.25177 , 1.13 , 1.734886], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 2.706187], [ 2.982142, 10.223478, 7.525549], [ 4.752927, 11.635139, 9.145517], [ 4.361189, 8.948196, 7.862798], [ 2.043874, 5.331582, 4.50407 ]], dtype=float32) - FSNT_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Net solar flux at top of model
- basename :
- FSNT
array([[1.499637e+00, 8.035751e+00, 1.407805e+01], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 7.519877e-04], [8.538224e-01, 9.882076e+00, 1.492072e+01], [1.478343e+01, 3.221646e+01, 3.289617e+01], [2.228963e+01, 3.931536e+01, 4.060257e+01], [2.208170e+01, 3.498952e+01, 3.811206e+01], [1.322824e+01, 2.417063e+01, 2.789650e+01]], dtype=float32) - FSNTOAC_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Clearsky net solar flux at top of atmosphere
- basename :
- FSNTOAC
array([[1.893185e+00, 1.000114e+01, 1.647750e+01], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 1.582047e-03], [8.534737e-01, 1.162448e+01, 1.953361e+01], [2.352954e+01, 4.812628e+01, 4.603608e+01], [4.045189e+01, 6.302671e+01, 5.838688e+01], [3.955408e+01, 5.925581e+01, 5.634935e+01], [2.133899e+01, 3.874036e+01, 4.045929e+01]], dtype=float32) - FSUTOA_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Upwelling solar flux at top of atmosphere
- basename :
- FSUTOA
array([[1.282829e+00, 7.504503e+00, 1.667826e+01], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 3.706536e-03], [1.023316e+00, 1.763088e+01, 2.214322e+01], [2.039597e+01, 4.663649e+01, 5.976355e+01], [3.838982e+01, 6.368174e+01, 7.927594e+01], [3.756379e+01, 6.294092e+01, 7.871978e+01], [1.890298e+01, 3.980881e+01, 5.581633e+01]], dtype=float32) - GCLDLWP_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/m2
- long_name :
- Grid-box cloud water path
- basename :
- GCLDLWP
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - ICE_ICLD_VISTAU_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- Sampling_Sequence :
- rad_lwsw
- units :
- 1
- long_name :
- Ice in-cloud extinction visible sw optical depth
- basename :
- ICE_ICLD_VISTAU
array([[[ 0., 0., 0.], [ 0., 0., 0.], ..., [ 0., 0., 0.], [ 0., 0., 0.]], [[nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan]], ..., [[ 0., 0., 0.], [ 0., 0., 0.], ..., [ 0., 0., 0.], [ 0., 0., 0.]], [[ 0., 0., 0.], [ 0., 0., 0.], ..., [ 0., 0., 0.], [ 0., 0., 0.]]], dtype=float32) - ICIMR_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Prognostic in-cloud ice mixing ratio
- basename :
- ICIMR
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - ICIMRST_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Prognostic in-stratus ice mixing ratio
- basename :
- ICIMRST
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - ICINC_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- m-3
- long_name :
- Prognostic in-cloud ice number conc
- basename :
- ICINC
array([[[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [5.448322e+00, 5.457217e-01, 8.021637e-01], [1.442298e+00, 4.038450e-01, 8.900693e-02]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [4.028392e+00, 4.741270e-01, 1.208132e+00], [1.009633e+00, 3.532165e-01, 1.315138e-01]], ..., [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [7.649843e-04, 1.922285e-03, 3.762565e-02], [3.980516e-04, 1.921996e-03, 3.325015e-02]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [6.653163e-04, 1.198170e-01, 1.751801e-02], [3.214641e-04, 8.615315e-03, 9.678490e-03]]], dtype=float32) - ICLDIWP_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/m2
- long_name :
- In-cloud ice water path
- basename :
- ICLDIWP
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - ICLDTWP_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/m2
- long_name :
- In-cloud cloud total water path (liquid and ice)
- basename :
- ICLDTWP
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - ICWMRST_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Prognostic in-stratus water mixing ratio
- basename :
- ICWMRST
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - ICWNC_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- m-3
- long_name :
- Prognostic in-cloud water number conc
- basename :
- ICWNC
array([[[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [1.342779e-08, 1.330742e-08, 3.466886e-10], [1.347290e-08, 1.335249e-08, 2.583221e-09]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [1.342714e-08, 1.330773e-08, 2.561702e-10], [1.347242e-08, 1.335273e-08, 2.096554e-09]], ..., [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [1.053975e-08, 2.054981e-09, 1.323694e-08], [1.302241e-08, 4.393191e-09, 1.328364e-08]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [1.062194e-08, 5.128133e-09, 1.318194e-08], [1.302227e-08, 1.118312e-08, 1.322817e-08]]], dtype=float32) - IWC_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/m3
- long_name :
- Grid box average ice water content
- basename :
- IWC
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - LIQ_ICLD_VISTAU_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- Sampling_Sequence :
- rad_lwsw
- units :
- 1
- long_name :
- Liquid in-cloud extinction visible sw optical depth
- basename :
- LIQ_ICLD_VISTAU
array([[[ 0., 0., 0.], [ 0., 0., 0.], ..., [ 0., 0., 0.], [ 0., 0., 0.]], [[nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan]], ..., [[ 0., 0., 0.], [ 0., 0., 0.], ..., [ 0., 0., 0.], [ 0., 0., 0.]], [[ 0., 0., 0.], [ 0., 0., 0.], ..., [ 0., 0., 0.], [ 0., 0., 0.]]], dtype=float32) - LWC_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/m3
- long_name :
- Grid box average liquid water content
- basename :
- LWC
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - LWCF_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Longwave cloud forcing
- basename :
- LWCF
array([[21.720066, 26.088118, 21.480127], [21.628384, 25.18713 , 20.91648 ], [22.117128, 24.73137 , 20.36388 ], [22.89166 , 24.308146, 19.924183], [23.587181, 23.923824, 19.504282], [28.35875 , 23.812637, 18.908743], [35.497795, 23.961866, 18.399971], [40.291237, 24.312006, 18.063877], [43.64851 , 24.454327, 17.741922], [45.169262, 25.282734, 17.368832], [50.111767, 27.606243, 17.074776], [54.338596, 31.096115, 14.200049], [55.69818 , 33.043224, 14.64996 ], [57.89926 , 36.51449 , 16.067335], [58.376057, 37.259144, 18.139975], [67.68696 , 35.88557 , 19.05946 ], [64.31541 , 54.427338, 31.430033], [66.07005 , 60.166695, 46.220898], [59.75964 , 62.18478 , 46.8051 ], [54.95062 , 58.25685 , 43.912178], [58.839977, 55.005943, 43.295322], [64.75803 , 51.796204, 42.121468], [61.02838 , 59.613667, 48.58198 ], [53.625328, 61.23687 , 52.313988]], dtype=float32) - NSNOW_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- m-3
- long_name :
- Diagnostic grid-mean snow number conc
- basename :
- NSNOW
array([[[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [9.570829e-03, 5.187288e+01, 3.837398e+02], [9.381848e-03, 4.119793e+01, 2.355800e+02]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [6.654274e-03, 5.532943e+01, 3.729627e+02], [6.503611e-03, 4.358483e+01, 2.237049e+02]], ..., [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [2.661725e+02, 3.028694e+02, 1.563086e+01], [2.396860e+02, 2.792774e+02, 1.118287e+01]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [2.230626e+02, 5.142866e+02, 1.890629e+02], [2.040495e+02, 4.762264e+02, 1.632102e+02]]], dtype=float32) - NUMICE_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- 1/kg
- long_name :
- Grid box averaged cloud ice number
- basename :
- NUMICE
array([[[1.000000e-12, 1.000000e-12, 1.000000e-12], [1.000000e-12, 1.000000e-12, 1.000000e-12], ..., [4.057496e-04, 4.100884e-05, 2.313787e-03], [1.070518e-04, 3.024492e-05, 3.445580e-05]], [[1.000000e-12, 1.000000e-12, 1.000000e-12], [1.000000e-12, 1.000000e-12, 1.000000e-12], ..., [3.000185e-04, 3.562794e-05, 4.716129e-03], [7.494078e-05, 2.645276e-05, 6.272856e-05]], ..., [[1.000000e-12, 1.000000e-12, 1.000000e-12], [1.000000e-12, 1.000000e-12, 1.000000e-12], ..., [7.258085e-08, 9.354276e-07, 2.842474e-06], [3.056667e-08, 4.374943e-07, 2.503090e-06]], [[1.000000e-12, 1.000000e-12, 1.000000e-12], [1.000000e-12, 1.000000e-12, 1.000000e-12], ..., [6.263608e-08, 2.336464e-05, 1.328940e-06], [2.468573e-08, 7.703853e-07, 7.316575e-07]]], dtype=float32) - NUMIMM10sBC_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- #/m3
- long_name :
- Ice Number Concentration due to imm freezing by bc in Mixed Clouds during 10-s period
- basename :
- NUMIMM10sBC
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - NUMIMM10sDST_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- #/m3
- long_name :
- Ice Number Concentration due to imm freezing by dst in Mixed Clouds during 10-s period
- basename :
- NUMIMM10sDST
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - NUMLIQ_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- 1/kg
- long_name :
- Grid box averaged cloud liquid number
- basename :
- NUMLIQ
array([[[1.e-12, 1.e-12, 1.e-12], [1.e-12, 1.e-12, 1.e-12], ..., [1.e-12, 1.e-12, 1.e-12], [1.e-12, 1.e-12, 1.e-12]], [[1.e-12, 1.e-12, 1.e-12], [1.e-12, 1.e-12, 1.e-12], ..., [1.e-12, 1.e-12, 1.e-12], [1.e-12, 1.e-12, 1.e-12]], ..., [[1.e-12, 1.e-12, 1.e-12], [1.e-12, 1.e-12, 1.e-12], ..., [1.e-12, 1.e-12, 1.e-12], [1.e-12, 1.e-12, 1.e-12]], [[1.e-12, 1.e-12, 1.e-12], [1.e-12, 1.e-12, 1.e-12], ..., [1.e-12, 1.e-12, 1.e-12], [1.e-12, 1.e-12, 1.e-12]]], dtype=float32) - NUMRAI_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- 1/kg
- long_name :
- Grid box averaged rain number
- basename :
- NUMRAI
array([[[1.000000e-12, 1.000000e-12, 1.000000e-12], [1.000000e-12, 1.000000e-12, 1.000000e-12], ..., [3.612901e+03, 3.996770e+03, 5.855105e+03], [1.444218e+03, 2.297924e+03, 3.641523e+03]], [[1.000000e-12, 1.000000e-12, 1.000000e-12], [1.000000e-12, 1.000000e-12, 1.000000e-12], ..., [3.006462e+03, 6.091466e+03, 5.424079e+03], [1.184377e+03, 1.720089e+03, 3.536481e+03]], ..., [[1.000000e-12, 1.000000e-12, 1.000000e-12], [1.000000e-12, 1.000000e-12, 1.000000e-12], ..., [3.956565e+03, 1.687498e+03, 7.568622e+00], [2.000783e+03, 9.052639e+02, 3.386011e+00]], [[1.000000e-12, 1.000000e-12, 1.000000e-12], [1.000000e-12, 1.000000e-12, 1.000000e-12], ..., [5.016835e+03, 2.895299e+03, 6.543821e+02], [2.508044e+03, 1.606233e+03, 3.307251e+02]]], dtype=float32) - NUMSNO_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- 1/kg
- long_name :
- Grid box averaged snow number
- basename :
- NUMSNO
array([[[1.000000e-12, 1.000000e-12, 1.000000e-12], [1.000000e-12, 1.000000e-12, 1.000000e-12], ..., [8.528994e-03, 2.560251e+01, 3.076804e+02], [8.435516e-03, 2.488488e+01, 1.354407e+02]], [[1.000000e-12, 1.000000e-12, 1.000000e-12], [1.000000e-12, 1.000000e-12, 1.000000e-12], ..., [5.993129e-03, 2.640857e+01, 3.036957e+02], [5.876542e-03, 2.574241e+01, 1.328176e+02]], ..., [[1.000000e-12, 1.000000e-12, 1.000000e-12], [1.000000e-12, 1.000000e-12, 1.000000e-12], ..., [1.634564e+02, 2.229083e+02, 7.963551e+00], [1.362070e+02, 1.922407e+02, 4.728848e+00]], [[1.000000e-12, 1.000000e-12, 1.000000e-12], [1.000000e-12, 1.000000e-12, 1.000000e-12], ..., [1.417755e+02, 3.540432e+02, 1.457527e+02], [1.211202e+02, 3.068614e+02, 1.140106e+02]]], dtype=float32) - OMEGA_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- Pa/s
- long_name :
- Vertical velocity (pressure)
- basename :
- OMEGA
array([[[ 9.201306e-05, 4.284060e-05, 3.211038e-05], [ 2.009390e-04, 3.669394e-05, -2.996684e-06], ..., [ 1.647879e-02, -3.492018e-02, -5.449503e-02], [ 1.072478e-02, -3.160384e-02, -3.811219e-02]], [[ 5.222417e-05, -1.419010e-06, -2.327440e-06], [ 1.237535e-04, -2.363608e-05, -3.579292e-05], ..., [ 1.316748e-02, -3.795349e-02, -5.490819e-02], [ 6.956419e-03, -3.499321e-02, -3.812005e-02]], ..., [[ 4.828516e-05, 3.624342e-05, 2.737288e-05], [ 1.362425e-04, 1.091857e-04, 1.158487e-04], ..., [-6.342716e-02, -3.159204e-02, 6.348663e-02], [-4.718038e-02, -2.610605e-02, 6.219332e-02]], [[ 3.500340e-05, 6.287876e-05, 5.858911e-05], [ 1.075895e-04, 1.494508e-04, 1.520548e-04], ..., [-4.872682e-02, -3.647901e-02, 4.960513e-02], [-3.418122e-02, -2.608609e-02, 5.495125e-02]]], dtype=float32) - PRAO_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg/s
- long_name :
- Accretion of cloud water by rain
- basename :
- PRAO
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - PRCO_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg/s
- long_name :
- Autoconversion of cloud water
- basename :
- PRCO
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - PRECC_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- units :
- m/s
- long_name :
- Convective precipitation rate (liq + ice)
- basename :
- PRECC
array([[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], dtype=float32) - PRECL_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- units :
- m/s
- long_name :
- Large-scale (stable) precipitation rate (liq + ice)
- basename :
- PRECL
array([[5.929956e-09, 1.951259e-09, 2.927100e-09], [4.444310e-09, 1.951826e-09, 3.176058e-09], [9.724466e-10, 1.645675e-09, 3.287522e-09], [8.169432e-10, 7.018801e-10, 1.840671e-09], [9.044606e-10, 1.633907e-09, 3.347234e-09], [1.092911e-09, 1.053148e-09, 3.183331e-09], [2.495869e-09, 6.667579e-10, 2.872639e-09], [4.177466e-09, 6.274303e-10, 1.821267e-09], [7.185577e-09, 8.915065e-10, 1.659624e-09], [1.202458e-08, 1.429613e-09, 2.057818e-09], [1.604454e-08, 1.263365e-09, 1.856049e-09], [2.151796e-08, 1.187000e-09, 1.999515e-09], [3.307302e-08, 3.130312e-09, 1.130926e-09], [4.741196e-08, 5.019779e-09, 2.482905e-09], [6.060783e-08, 8.877103e-09, 5.653991e-09], [7.245784e-08, 1.241478e-08, 8.411678e-09], [8.822788e-08, 1.783736e-08, 1.259261e-08], [1.242271e-07, 2.679551e-08, 1.944385e-08], [1.431587e-07, 3.737021e-08, 2.676024e-08], [1.535562e-07, 5.840652e-08, 3.991724e-08], [1.413443e-07, 6.952206e-08, 4.998965e-08], [1.252556e-07, 7.671156e-08, 5.860462e-08], [1.049210e-07, 9.030627e-08, 7.079801e-08], [7.996199e-08, 1.220762e-07, 8.100228e-08]], dtype=float32) - PRECT_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- units :
- m/s
- long_name :
- Total (convective and large-scale) precipitation rate (liq + ice)
- basename :
- PRECT
array([[5.929956e-09, 1.951259e-09, 2.927100e-09], [4.444310e-09, 1.951826e-09, 3.176058e-09], [9.724466e-10, 1.645675e-09, 3.287522e-09], [8.169432e-10, 7.018801e-10, 1.840671e-09], [9.044606e-10, 1.633907e-09, 3.347234e-09], [1.092911e-09, 1.053148e-09, 3.183331e-09], [2.495869e-09, 6.667579e-10, 2.872639e-09], [4.177466e-09, 6.274303e-10, 1.821267e-09], [7.185577e-09, 8.915065e-10, 1.659624e-09], [1.202458e-08, 1.429613e-09, 2.057818e-09], [1.604454e-08, 1.263365e-09, 1.856049e-09], [2.151796e-08, 1.187000e-09, 1.999515e-09], [3.307302e-08, 3.130312e-09, 1.130926e-09], [4.741196e-08, 5.019779e-09, 2.482905e-09], [6.060783e-08, 8.877103e-09, 5.653991e-09], [7.245784e-08, 1.241478e-08, 8.411678e-09], [8.822788e-08, 1.783736e-08, 1.259261e-08], [1.242271e-07, 2.679551e-08, 1.944385e-08], [1.431587e-07, 3.737021e-08, 2.676024e-08], [1.535562e-07, 5.840652e-08, 3.991724e-08], [1.413443e-07, 6.952206e-08, 4.998965e-08], [1.252556e-07, 7.671156e-08, 5.860462e-08], [1.049210e-07, 9.030627e-08, 7.079801e-08], [7.996199e-08, 1.220762e-07, 8.100228e-08]], dtype=float32) - PS_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- units :
- Pa
- long_name :
- Surface pressure
- basename :
- PS
array([[103446.984, 103024.234, 102308.47 ], [103478.01 , 103074.67 , 102363.61 ], [103508.875, 103130.77 , 102424.13 ], [103528.22 , 103179.96 , 102479.14 ], [103528.875, 103212.375, 102514.34 ], [103511.91 , 103237.81 , 102545.625], [103491.79 , 103260.92 , 102571.195], [103459.57 , 103280.01 , 102593.38 ], [103415.914, 103296.08 , 102613.805], [103372.62 , 103320.23 , 102644.82 ], [103303.18 , 103323.2 , 102655.914], [103214.25 , 103312.4 , 102656.41 ], [103113.4 , 103288.53 , 102648.57 ], [102988.73 , 103240.195, 102616.97 ], [102853.82 , 103187.5 , 102580.79 ], [102712.39 , 103118.41 , 102529.76 ], [102578.14 , 103054.86 , 102484.74 ], [102436.42 , 102969.05 , 102419.984], [102305.47 , 102868.5 , 102342.445], [102190.47 , 102768.414, 102259.03 ], [102102.234, 102665.336, 102175.945], [102008.81 , 102531.46 , 102064.64 ], [101924.33 , 102378.85 , 101931.945], [101875.07 , 102241.84 , 101811.42 ]], dtype=float32) - PSL_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- units :
- Pa
- long_name :
- Sea level pressure
- basename :
- PSL
array([[103447.58 , 103290.86 , 103336.76 ], [103478.61 , 103341.305, 103392.1 ], [103509.48 , 103397.43 , 103452.84 ], [103528.82 , 103446.63 , 103508.08 ], [103529.48 , 103479.04 , 103543.36 ], [103512.5 , 103504.46 , 103574.766], [103492.39 , 103527.57 , 103600.555], [103460.164, 103546.64 , 103622.97 ], [103416.51 , 103562.7 , 103643.79 ], [103373.21 , 103586.91 , 103675.45 ], [103303.77 , 103589.875, 103687.31 ], [103214.84 , 103579.02 , 103690.61 ], [103113.984, 103555.08 , 103686.664], [102989.31 , 103506.64 , 103658.15 ], [102854.41 , 103453.875, 103622.88 ], [102712.98 , 103384.695, 103571.61 ], [102578.73 , 103321.05 , 103525.44 ], [102437. , 103235.07 , 103458.51 ], [102306.055, 103134.266, 103378.945], [102191.05 , 103033.84 , 103293.39 ], [102102.81 , 102930.336, 103208.016], [102009.39 , 102795.836, 103093.47 ], [101924.91 , 102642.43 , 102956.85 ], [101875.65 , 102504.57 , 102832.04 ]], dtype=float32) - Q_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Specific humidity
- basename :
- Q
array([[[2.128108e-06, 2.163354e-06, 2.190013e-06], [2.154065e-06, 2.141479e-06, 2.143832e-06], ..., [1.905315e-03, 1.885452e-03, 1.886655e-03], [1.912382e-03, 1.889003e-03, 1.898013e-03]], [[2.139954e-06, 2.174797e-06, 2.205221e-06], [2.152838e-06, 2.151143e-06, 2.158222e-06], ..., [1.903464e-03, 1.934436e-03, 1.918999e-03], [1.910334e-03, 1.937764e-03, 1.928606e-03]], ..., [[2.159604e-06, 2.166671e-06, 2.177318e-06], [2.166007e-06, 2.166609e-06, 2.164300e-06], ..., [2.949869e-03, 2.514976e-03, 2.045970e-03], [2.954819e-03, 2.519611e-03, 2.049868e-03]], [[2.170608e-06, 2.163529e-06, 2.172495e-06], [2.166615e-06, 2.169783e-06, 2.166471e-06], ..., [2.928583e-03, 2.634722e-03, 2.203714e-03], [2.933594e-03, 2.639771e-03, 2.206351e-03]]], dtype=float32) - QRAIN_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Diagnostic grid-mean rain mixing ratio
- basename :
- QRAIN
array([[[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [1.914412e-06, 4.179978e-07, 5.091842e-07], [1.479660e-06, 2.502543e-07, 2.847929e-07]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [1.473782e-06, 4.627196e-07, 6.069883e-07], [1.140054e-06, 2.260730e-07, 3.583910e-07]], ..., [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [9.983011e-06, 3.336476e-06, 9.713277e-08], [9.373496e-06, 3.116626e-06, 8.232237e-08]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [1.009182e-05, 6.681274e-06, 1.989750e-06], [9.327486e-06, 6.275529e-06, 1.778090e-06]]], dtype=float32) - QSNOW_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Diagnostic grid-mean snow mixing ratio
- basename :
- QSNOW
array([[[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [1.981317e-09, 1.436742e-06, 4.219262e-06], [2.141916e-09, 1.251544e-06, 3.019975e-06]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [1.645345e-09, 1.575783e-06, 4.382458e-06], [1.820044e-09, 1.352032e-06, 3.089148e-06]], ..., [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [6.644947e-05, 6.550150e-05, 5.477341e-05], [6.654794e-05, 6.513305e-05, 5.465002e-05]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [4.896725e-05, 8.476641e-05, 5.790207e-05], [4.889019e-05, 8.437818e-05, 5.788096e-05]]], dtype=float32) - QVRES_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg/s
- long_name :
- Rate of residual condensation term
- basename :
- QVRES
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - RAINQM_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Grid box averaged rain amount
- basename :
- RAINQM
array([[[1.276701e-24, 1.285470e-24, 1.298394e-24], [1.232930e-24, 1.238952e-24, 1.251827e-24], ..., [7.041990e-06, 1.004498e-07, 3.580454e-07], [6.280679e-06, 1.573069e-07, 1.948113e-07]], [[1.263375e-24, 1.334355e-24, 1.355425e-24], [1.250935e-24, 1.273889e-24, 1.287261e-24], ..., [5.542766e-06, 1.911733e-07, 4.213022e-07], [4.918394e-06, 4.323054e-08, 2.384287e-07]], ..., [[1.169955e-24, 1.205623e-24, 1.241814e-24], [1.125649e-24, 1.128782e-24, 1.149575e-24], ..., [8.455470e-06, 3.066729e-06, 4.737110e-08], [7.242728e-06, 2.678074e-06, 3.668050e-08]], [[1.131833e-24, 1.164937e-24, 1.202065e-24], [1.098900e-24, 1.130117e-24, 1.164864e-24], ..., [8.983904e-06, 5.998549e-06, 1.833039e-06], [7.907805e-06, 5.298977e-06, 1.532744e-06]]], dtype=float32) - RELHUM_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- percent
- long_name :
- Relative humidity
- basename :
- RELHUM
array([[[2.128108e-04, 2.163354e-04, 2.190013e-04], [2.154065e-04, 2.141479e-04, 2.143832e-04], ..., [8.561469e+01, 7.578206e+01, 8.357990e+01], [8.373080e+01, 7.405994e+01, 8.256472e+01]], [[2.139954e-04, 2.174797e-04, 2.205221e-04], [2.152838e-04, 2.151143e-04, 2.158222e-04], ..., [8.490095e+01, 7.702303e+01, 8.437074e+01], [8.306506e+01, 7.522659e+01, 8.333339e+01]], ..., [[2.159604e-04, 2.166671e-04, 2.177318e-04], [2.166007e-04, 2.166609e-04, 2.164300e-04], ..., [8.678175e+01, 8.988383e+01, 9.147659e+01], [8.501234e+01, 8.780931e+01, 8.960609e+01]], [[2.170608e-04, 2.163529e-04, 2.172495e-04], [2.166615e-04, 2.169783e-04, 2.166471e-04], ..., [8.695348e+01, 9.031312e+01, 9.218178e+01], [8.515396e+01, 8.832088e+01, 9.018970e+01]]], dtype=float32) - RERCLD_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- m
- long_name :
- Diagnostic effective radius of Liquid Cloud and Rain
- basename :
- RERCLD
array([[[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [5.193589e-05, 4.140491e-05, 4.366448e-05], [5.910731e-05, 3.796657e-05, 4.176056e-05]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [5.536053e-05, 3.959521e-05, 4.726124e-05], [6.304363e-05, 3.818220e-05, 4.562473e-05]], ..., [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [5.218072e-05, 1.006312e-04, 1.728775e-04], [5.556297e-05, 1.094717e-04, 1.975507e-04]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [5.066645e-05, 7.736035e-05, 1.007705e-04], [5.494227e-05, 6.701393e-05, 1.177945e-04]]], dtype=float32) - RHCFMIP_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- percent
- long_name :
- Relative humidity with respect to water above 273 K, ice below 273 K
- basename :
- RHCFMIP
array([[[8.829004e-06, 6.153696e-06, 5.531151e-06], [1.222711e-05, 8.695690e-06, 7.924716e-06], ..., [8.946259e+01, 7.874077e+01, 8.732493e+01], [8.734685e+01, 7.679969e+01, 8.615706e+01]], [[8.436945e-06, 5.433798e-06, 4.894741e-06], [1.040160e-05, 7.350379e-06, 6.788883e-06], ..., [8.868682e+01, 7.996557e+01, 8.811757e+01], [8.660671e+01, 7.797271e+01, 8.691586e+01]], ..., [[1.302890e-05, 1.000152e-05, 9.640508e-06], [2.833140e-05, 2.426895e-05, 2.279303e-05], ..., [8.791240e+01, 9.267970e+01, 9.562117e+01], [8.580856e+01, 9.034975e+01, 9.355365e+01]], [[1.308286e-05, 9.493470e-06, 8.739089e-06], [2.396412e-05, 1.944842e-05, 1.815056e-05], ..., [8.818503e+01, 9.283851e+01, 9.603873e+01], [8.606655e+01, 9.052531e+01, 9.385751e+01]]], dtype=float32) - RHI_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- percent
- long_name :
- Relative humidity with respect to ice
- basename :
- RHI
array([[[8.829004e-06, 6.153696e-06, 5.531151e-06], [1.222711e-05, 8.695690e-06, 7.924716e-06], ..., [8.946259e+01, 7.874077e+01, 8.732493e+01], [8.734685e+01, 7.679969e+01, 8.615706e+01]], [[8.436945e-06, 5.433798e-06, 4.894741e-06], [1.040160e-05, 7.350379e-06, 6.788883e-06], ..., [8.868682e+01, 7.996557e+01, 8.811757e+01], [8.660671e+01, 7.797271e+01, 8.691586e+01]], ..., [[1.302890e-05, 1.000152e-05, 9.640508e-06], [2.833140e-05, 2.426895e-05, 2.279303e-05], ..., [8.791240e+01, 9.267970e+01, 9.562117e+01], [8.580856e+01, 9.034975e+01, 9.355365e+01]], [[1.308286e-05, 9.493470e-06, 8.739089e-06], [2.396412e-05, 1.944842e-05, 1.815056e-05], ..., [8.818503e+01, 9.283851e+01, 9.603873e+01], [8.606655e+01, 9.052531e+01, 9.385751e+01]]], dtype=float32) - RHW_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- percent
- long_name :
- Relative humidity with respect to liquid
- basename :
- RHW
array([[[2.128108e-04, 2.163354e-04, 2.190013e-04], [2.154065e-04, 2.141479e-04, 2.143832e-04], ..., [8.394084e+01, 7.483093e+01, 8.196519e+01], [8.225536e+01, 7.324888e+01, 8.109003e+01]], [[2.139954e-04, 2.174797e-04, 2.205221e-04], [2.152838e-04, 2.151143e-04, 2.158222e-04], ..., [8.328826e+01, 7.608734e+01, 8.278838e+01], [8.162962e+01, 7.445930e+01, 8.187582e+01]], ..., [[2.159604e-04, 2.166671e-04, 2.177318e-04], [2.166007e-04, 2.166609e-04, 2.164300e-04], ..., [8.674396e+01, 8.927917e+01, 8.961723e+01], [8.497119e+01, 8.735542e+01, 8.796146e+01]], [[2.170608e-04, 2.163529e-04, 2.172495e-04], [2.166615e-04, 2.169783e-04, 2.166471e-04], ..., [8.690346e+01, 8.987926e+01, 9.070859e+01], [8.512042e+01, 8.796225e+01, 8.893913e+01]]], dtype=float32) - SNOWQM_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Grid box averaged snow amount
- basename :
- SNOWQM
array([[[2.227845e-24, 2.194918e-24, 2.211645e-24], [2.293272e-24, 2.209818e-24, 2.211099e-24], ..., [2.208568e-09, 1.051507e-06, 4.354991e-06], [2.413437e-09, 1.036379e-06, 2.530505e-06]], [[2.253171e-24, 2.222106e-24, 2.230408e-24], [2.309606e-24, 2.244894e-24, 2.250696e-24], ..., [1.832344e-09, 1.136848e-06, 4.531147e-06], [2.016452e-09, 1.113002e-06, 2.613673e-06]], ..., [[2.214561e-24, 2.205818e-24, 2.206141e-24], [2.319602e-24, 2.317127e-24, 2.304383e-24], ..., [6.678556e-05, 6.645532e-05, 5.432707e-05], [6.737564e-05, 6.627461e-05, 5.435228e-05]], [[2.237489e-24, 2.223133e-24, 2.228757e-24], [2.322820e-24, 2.307803e-24, 2.279569e-24], ..., [4.989577e-05, 8.539884e-05, 6.000307e-05], [5.028994e-05, 8.539146e-05, 5.997338e-05]]], dtype=float32) - SNOW_ICLD_VISTAU_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- Sampling_Sequence :
- rad_lwsw
- units :
- 1
- long_name :
- Snow in-cloud extinction visible sw optical depth
- basename :
- SNOW_ICLD_VISTAU
array([[[0. , 0. , 0. ], [0. , 0. , 0. ], ..., [0. , 0.004579, 0.017905], [0. , 0.001852, 0.005498]], [[ nan, nan, nan], [ nan, nan, nan], ..., [ nan, nan, nan], [ nan, nan, nan]], ..., [[0. , 0. , 0. ], [0. , 0. , 0. ], ..., [0.095467, 0.066042, 0.057306], [0.041211, 0.029195, 0.026611]], [[0. , 0. , 0. ], [0. , 0. , 0. ], ..., [0.079415, 0.121213, 0.052388], [0.034787, 0.053417, 0.024389]]], dtype=float32) - SOLIN_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Solar insolation
- basename :
- SOLIN
array([[2.817350e+00, 1.562921e+01, 3.086798e+01], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 5.353549e-03], [1.904548e+00, 2.762606e+01, 3.718538e+01], [3.530029e+01, 7.899471e+01, 9.280216e+01], [6.081453e+01, 1.031428e+02, 1.200240e+02], [5.978024e+01, 9.807334e+01, 1.169774e+02], [3.224912e+01, 6.411356e+01, 8.385178e+01]], dtype=float32) - SWCF_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Shortwave cloud forcing
- basename :
- SWCF
array([[-3.586587e-01, -1.876408e+00, -2.287746e+00], [ 0.000000e+00, 0.000000e+00, 0.000000e+00], [ 0.000000e+00, 0.000000e+00, 0.000000e+00], [ 0.000000e+00, 0.000000e+00, 0.000000e+00], [ 0.000000e+00, 0.000000e+00, 0.000000e+00], [ 0.000000e+00, 0.000000e+00, 0.000000e+00], [ 0.000000e+00, 0.000000e+00, 0.000000e+00], [ 0.000000e+00, 0.000000e+00, 0.000000e+00], [ 0.000000e+00, 0.000000e+00, 0.000000e+00], [ 0.000000e+00, 0.000000e+00, 0.000000e+00], [ 0.000000e+00, 0.000000e+00, 0.000000e+00], [ 0.000000e+00, 0.000000e+00, 0.000000e+00], [ 0.000000e+00, 0.000000e+00, 0.000000e+00], [ 0.000000e+00, 0.000000e+00, 0.000000e+00], [ 0.000000e+00, 0.000000e+00, 0.000000e+00], [ 0.000000e+00, 0.000000e+00, 0.000000e+00], [ 0.000000e+00, 0.000000e+00, 0.000000e+00], [ 0.000000e+00, 0.000000e+00, 0.000000e+00], [ 0.000000e+00, 0.000000e+00, 6.497469e-05], [ 2.776120e-02, -1.629261e+00, -4.491395e+00], [-8.625181e+00, -1.576794e+01, -1.299732e+01], [-1.802710e+01, -2.356553e+01, -1.763868e+01], [-1.733754e+01, -2.412324e+01, -1.809159e+01], [-7.992801e+00, -1.443550e+01, -1.242370e+01]], dtype=float32) - T_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- K
- long_name :
- Temperature
- basename :
- T
array([[[264.73633, 269.11035, 270.51874], [265.60776, 269.5085 , 270.6233 ], ..., [266.61496, 267.9273 , 266.65262], [266.9897 , 268.29642, 266.93283]], [[265.3187 , 270.64722, 272.0655 ], [267.46976, 271.5637 , 272.5604 ], ..., [266.70782, 268.0522 , 266.75085], [267.0793 , 268.423 , 267.0227 ]], ..., [[260.53613, 263.5252 , 263.99664], [256.36392, 258.0316 , 258.70105], ..., [271.79153, 269.3201 , 266.4981 ], [272.16 , 269.69852, 266.82767]], [[260.5466 , 264.09888, 265.0888 ], [258.16855, 260.46756, 261.2148 ], ..., [271.66174, 269.8332 , 267.29352], [272.03137, 270.21118, 267.6298 ]]], dtype=float32) - TGCLDCWP_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- units :
- kg/m2
- long_name :
- Total grid-box cloud water path (liquid and ice)
- basename :
- TGCLDCWP
array([[0.181146, 0.228208, 0.155753], [0.174493, 0.224488, 0.160122], [0.168044, 0.213729, 0.158764], [0.162022, 0.2066 , 0.149719], [0.15009 , 0.200088, 0.133257], [0.136189, 0.190127, 0.119104], [0.119106, 0.175186, 0.102689], [0.107121, 0.151274, 0.083715], [0.093964, 0.116704, 0.065929], [0.085536, 0.07963 , 0.046929], [0.102502, 0.047249, 0.020328], [0.120582, 0.023668, 0.00812 ], [0.153632, 0.017285, 0.004612], [0.182647, 0.014937, 0.005808], [0.202993, 0.009571, 0.006517], [0.221541, 0.017487, 0.0064 ], [0.220088, 0.037908, 0.017773], [0.222036, 0.046242, 0.036391], [0.183962, 0.057164, 0.044538], [0.186125, 0.055711, 0.056541], [0.235994, 0.079146, 0.06788 ], [0.278998, 0.130476, 0.09562 ], [0.310765, 0.188159, 0.148169], [0.322842, 0.202387, 0.192566]], dtype=float32) - TGCLDIWP_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- units :
- kg/m2
- long_name :
- Total grid-box cloud ice water path
- basename :
- TGCLDIWP
array([[3.245805e-07, 1.319663e-03, 4.420833e-03], [1.121351e-04, 1.131896e-03, 3.595493e-03], [3.061806e-04, 1.051455e-03, 3.127915e-03], [5.213613e-04, 1.432166e-03, 3.332356e-03], [7.948484e-04, 1.134565e-03, 3.658754e-03], [2.781993e-03, 1.038114e-03, 3.234660e-03], [5.085572e-03, 1.278872e-03, 2.962676e-03], [6.825811e-03, 1.796853e-03, 2.820599e-03], [8.125892e-03, 2.257235e-03, 1.870526e-03], [8.128699e-03, 2.901913e-03, 1.524942e-03], [8.256746e-03, 3.425796e-03, 1.441408e-03], [9.568190e-03, 4.760813e-03, 2.560055e-03], [9.550201e-03, 6.151294e-03, 4.612403e-03], [9.772433e-03, 7.228503e-03, 5.807842e-03], [1.476581e-02, 7.067846e-03, 6.517095e-03], [1.753853e-02, 7.961069e-03, 6.399870e-03], [1.537306e-02, 9.860369e-03, 8.330411e-03], [4.012157e-02, 2.267269e-02, 1.383178e-02], [2.602716e-02, 3.594816e-02, 2.324953e-02], [1.518873e-02, 2.591780e-02, 2.619517e-02], [1.159262e-02, 2.194845e-02, 1.961996e-02], [1.204778e-02, 1.662401e-02, 1.493191e-02], [1.138118e-02, 2.047024e-02, 1.689687e-02], [1.463244e-02, 1.814569e-02, 1.427413e-02]], dtype=float32) - TGCLDLWP_155w_to_158w_70n_to_73n(time, ncol_155w_to_158w_70n_to_73n)float32...
- units :
- kg/m2
- long_name :
- Total grid-box cloud liquid water path
- basename :
- TGCLDLWP
array([[1.811461e-01, 2.268879e-01, 1.513323e-01], [1.743813e-01, 2.233557e-01, 1.565266e-01], [1.677379e-01, 2.126779e-01, 1.556358e-01], [1.615002e-01, 2.051682e-01, 1.463870e-01], [1.492950e-01, 1.989530e-01, 1.295982e-01], [1.334067e-01, 1.890886e-01, 1.158697e-01], [1.140208e-01, 1.739069e-01, 9.972633e-02], [1.002948e-01, 1.494768e-01, 8.089466e-02], [8.583828e-02, 1.144463e-01, 6.405845e-02], [7.740708e-02, 7.672817e-02, 4.540380e-02], [9.424561e-02, 4.382358e-02, 1.888703e-02], [1.110137e-01, 1.890768e-02, 5.559628e-03], [1.440815e-01, 1.113383e-02, 9.728568e-14], [1.728750e-01, 7.708400e-03, 0.000000e+00], [1.882267e-01, 2.502962e-03, 0.000000e+00], [2.040025e-01, 9.526066e-03, 0.000000e+00], [2.047153e-01, 2.804811e-02, 9.442569e-03], [1.819144e-01, 2.356928e-02, 2.255927e-02], [1.579353e-01, 2.121560e-02, 2.128874e-02], [1.709359e-01, 2.979312e-02, 3.034574e-02], [2.244010e-01, 5.719722e-02, 4.826012e-02], [2.669505e-01, 1.138521e-01, 8.068806e-02], [2.993836e-01, 1.676887e-01, 1.312716e-01], [3.082093e-01, 1.842413e-01, 1.782916e-01]], dtype=float32) - TOT_CLD_VISTAU_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- Sampling_Sequence :
- rad_lwsw
- units :
- 1
- long_name :
- Total gbx cloud extinction visible sw optical depth
- basename :
- TOT_CLD_VISTAU
array([[[0. , 0. , 0. ], [0. , 0. , 0. ], ..., [0. , 0.001145, 0.004476], [0. , 0.000463, 0.001374]], [[ nan, nan, nan], [ nan, nan, nan], ..., [ nan, nan, nan], [ nan, nan, nan]], ..., [[0. , 0. , 0. ], [0. , 0. , 0. ], ..., [0.023867, 0.01651 , 0.014326], [0.010303, 0.007299, 0.006653]], [[0. , 0. , 0. ], [0. , 0. , 0. ], ..., [0.019854, 0.030303, 0.013097], [0.008697, 0.013354, 0.006097]]], dtype=float32) - TOT_ICLD_VISTAU_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- Sampling_Sequence :
- rad_lwsw
- units :
- 1
- long_name :
- Total in-cloud extinction visible sw optical depth
- basename :
- TOT_ICLD_VISTAU
array([[[0. , 0. , 0. ], [0. , 0. , 0. ], ..., [0. , 0.004579, 0.017905], [0. , 0.001852, 0.005498]], [[ nan, nan, nan], [ nan, nan, nan], ..., [ nan, nan, nan], [ nan, nan, nan]], ..., [[0. , 0. , 0. ], [0. , 0. , 0. ], ..., [0.095467, 0.066042, 0.057306], [0.041211, 0.029195, 0.026611]], [[0. , 0. , 0. ], [0. , 0. , 0. ], ..., [0.079415, 0.121213, 0.052388], [0.034787, 0.053417, 0.024389]]], dtype=float32) - U_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- m/s
- long_name :
- Zonal wind
- basename :
- U
array([[[ 5.881633e+01, 4.243681e+01, 3.917900e+01], [ 6.178924e+01, 5.829353e+01, 5.912407e+01], ..., [ 2.731568e-01, -7.899590e-01, -2.962942e-01], [ 2.622272e-01, -7.117409e-01, -1.634923e-01]], [[ 5.869994e+01, 4.386864e+01, 4.062827e+01], [ 6.808576e+01, 6.810219e+01, 7.061655e+01], ..., [ 5.702493e-01, -6.746519e-01, -1.800306e-01], [ 5.336940e-01, -6.044235e-01, -7.190455e-02]], ..., [[ 7.215047e+01, 9.014555e+01, 9.705116e+01], [ 6.226543e+01, 7.580591e+01, 8.072163e+01], ..., [ 1.216581e+01, 1.248005e+01, 1.059074e+01], [ 1.107062e+01, 1.066450e+01, 7.900169e+00]], [[ 7.876751e+01, 9.854697e+01, 1.049941e+02], [ 7.208768e+01, 8.735288e+01, 9.151439e+01], ..., [ 1.146694e+01, 1.287841e+01, 1.124093e+01], [ 1.048071e+01, 1.097546e+01, 8.525111e+00]]], dtype=float32) - V_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- m/s
- long_name :
- Meridional wind
- basename :
- V
array([[[-4.427489, -4.090545, -2.853115], [17.39923 , 9.301125, 7.927942], ..., [-4.864511, -7.351467, -6.721163], [-4.542304, -6.798095, -4.848556]], [[-3.169981, -5.099699, -4.464707], [15.605526, 7.683027, 6.11841 ], ..., [-4.242459, -7.167257, -6.615004], [-3.962365, -6.623555, -4.723304]], ..., [[13.364609, 10.260371, 12.646693], [13.59661 , 9.956541, 10.788858], ..., [ 0.110032, 8.950782, 8.894286], [ 0.132383, 7.749557, 6.796473]], [[13.535832, 13.939431, 17.549086], [13.801663, 11.897252, 13.399579], ..., [-2.437408, 7.939637, 8.215149], [-2.196862, 6.865246, 6.369951]]], dtype=float32) - WSUB_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- m/s
- long_name :
- Diagnostic sub-grid vertical velocity
- basename :
- WSUB
array([[[0.2 , 0.2 , 0.2 ], [0.2 , 0.2 , 0.2 ], ..., [0.405321, 0.45578 , 0.307218], [0.299648, 0.406536, 0.32726 ]], [[0.2 , 0.2 , 0.2 ], [0.2 , 0.2 , 0.2 ], ..., [0.385158, 0.443574, 0.291971], [0.279941, 0.397065, 0.312101]], ..., [[0.2 , 0.2 , 0.2 ], [0.2 , 0.2 , 0.2 ], ..., [0.546208, 0.793169, 0.64544 ], [0.542441, 0.786902, 0.67821 ]], [[0.2 , 0.2 , 0.2 ], [0.2 , 0.2 , 0.2 ], ..., [0.521909, 0.779814, 0.67151 ], [0.51491 , 0.775581, 0.701208]]], dtype=float32) - WSUBI_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- m/s
- long_name :
- Diagnostic sub-grid vertical velocity for ice
- basename :
- WSUBI
array([[[0.001 , 0.001 , 0.001 ], [0.001 , 0.001 , 0.001 ], ..., [0.405321, 0.45578 , 0.307218], [0.299648, 0.406536, 0.32726 ]], [[0.001 , 0.001 , 0.001 ], [0.001 , 0.001 , 0.001 ], ..., [0.385158, 0.443574, 0.291971], [0.279941, 0.397065, 0.312101]], ..., [[0.001 , 0.001 , 0.001 ], [0.001 , 0.001 , 0.001 ], ..., [0.546208, 0.793169, 0.64544 ], [0.542441, 0.786902, 0.67821 ]], [[0.001 , 0.001 , 0.001 ], [0.001 , 0.001 , 0.001 ], ..., [0.521909, 0.779814, 0.67151 ], [0.51491 , 0.775581, 0.701208]]], dtype=float32) - Z3_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- m
- long_name :
- Geopotential Height (above sea level)
- basename :
- Z3
array([[[6.105669e+04, 6.126900e+04, 6.134579e+04], [5.806969e+04, 5.823539e+04, 5.829797e+04], ..., [4.905131e+01, 6.955544e+01, 1.272419e+02], [1.182723e+01, 3.214893e+01, 9.001710e+01]], [[6.104818e+04, 6.127150e+04, 6.135191e+04], [5.804741e+04, 5.821766e+04, 5.828446e+04], ..., [4.906803e+01, 6.957999e+01, 1.272601e+02], [1.183117e+01, 3.215487e+01, 9.002129e+01]], ..., [[6.051336e+04, 6.074804e+04, 6.084808e+04], [5.760207e+04, 5.781052e+04, 5.790414e+04], ..., [5.003326e+01, 6.983102e+01, 1.272225e+02], [1.206299e+01, 3.221536e+01, 9.001354e+01]], [[6.053251e+04, 6.079148e+04, 6.089944e+04], [5.761100e+04, 5.783702e+04, 5.793519e+04], ..., [5.000887e+01, 6.992888e+01, 1.273739e+02], [1.205716e+01, 3.223886e+01, 9.005006e+01]]], dtype=float32) - bc_a1_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- bc_a1 concentration
- basename :
- bc_a1
array([[[1.131937e-21, 1.506807e-21, 1.540672e-21], [1.730214e-21, 1.570911e-21, 1.417760e-21], ..., [1.784000e-12, 1.904676e-12, 2.229650e-12], [1.781611e-12, 1.902796e-12, 2.231422e-12]], [[1.188791e-21, 1.035877e-21, 1.108928e-21], [1.566421e-21, 1.129800e-21, 1.060017e-21], ..., [1.834711e-12, 1.860036e-12, 2.229998e-12], [1.832976e-12, 1.858097e-12, 2.231628e-12]], ..., [[4.827755e-22, 4.165199e-22, 4.280981e-22], [1.047410e-21, 1.018132e-21, 1.109655e-21], ..., [3.701343e-13, 2.499937e-12, 3.139591e-12], [3.699446e-13, 2.500649e-12, 3.148171e-12]], [[4.432855e-22, 4.160903e-22, 4.414517e-22], [8.495113e-22, 9.130608e-22, 9.904193e-22], ..., [3.203791e-13, 2.076417e-12, 2.557432e-12], [3.200552e-13, 2.078936e-12, 2.572068e-12]]], dtype=float32) - bc_a3_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- bc_a3 concentration
- basename :
- bc_a3
array([[[1.000000e-36, 1.008142e-36, 1.017086e-36], [1.000506e-36, 1.000000e-36, 1.000000e-36], ..., [8.098413e-14, 2.710656e-13, 4.004745e-13], [7.966464e-14, 2.718868e-13, 3.863113e-13]], [[1.000000e-36, 1.006386e-36, 1.023065e-36], [1.000197e-36, 1.000000e-36, 1.000000e-36], ..., [7.894408e-14, 3.358711e-13, 4.574602e-13], [7.787920e-14, 3.373081e-13, 4.383031e-13]], ..., [[1.002299e-36, 1.004569e-36, 1.008369e-36], [1.000143e-36, 1.000065e-36, 1.000000e-36], ..., [3.860658e-14, 7.091134e-14, 8.472153e-14], [3.824112e-14, 7.064009e-14, 8.503537e-14]], [[1.005624e-36, 1.001142e-36, 1.004832e-36], [1.000000e-36, 1.000637e-36, 1.000000e-36], ..., [5.097027e-14, 5.484377e-14, 6.306056e-14], [5.031199e-14, 5.486617e-14, 6.370704e-14]]], dtype=float32) - bc_a4_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- bc_a4 concentration
- basename :
- bc_a4
array([[[1.647139e-18, 1.585881e-18, 1.579548e-18], [1.725313e-18, 1.627352e-18, 1.636458e-18], ..., [2.009955e-12, 4.934976e-12, 8.079090e-12], [2.008279e-12, 4.951629e-12, 8.250519e-12]], [[1.656745e-18, 1.655805e-18, 1.660010e-18], [1.801513e-18, 1.700160e-18, 1.717517e-18], ..., [2.064600e-12, 5.214839e-12, 8.688733e-12], [2.063133e-12, 5.232793e-12, 8.909076e-12]], ..., [[1.298958e-18, 1.322708e-18, 1.411213e-18], [1.556754e-18, 1.568533e-18, 1.634346e-18], ..., [5.736931e-13, 5.085958e-12, 7.206440e-12], [5.743769e-13, 5.101794e-12, 7.256314e-12]], [[1.189183e-18, 1.232216e-18, 1.327401e-18], [1.406562e-18, 1.474589e-18, 1.571515e-18], ..., [6.466061e-13, 4.838702e-12, 6.078939e-12], [6.469959e-13, 4.854258e-12, 6.141941e-12]]], dtype=float32) - bc_c1_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- bc_c1 in cloud water
- basename :
- bc_c1
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - bc_c3_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- bc_c3 in cloud water
- basename :
- bc_c3
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - bc_c4_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- bc_c4 in cloud water
- basename :
- bc_c4
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - dei_cloud_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- micrometers
- long_name :
- ice radiative effective diameter in cloud
- basename :
- dei_cloud
array([[[ 27.262814, 27.262814, 27.262814], [ 27.262814, 27.262814, 27.262814], ..., [ 50. , 84.69093 , 94.32067 ], [ 50. , 109.03206 , 30.05722 ]], [[ 27.262814, 27.262814, 27.262814], [ 27.262814, 27.262814, 27.262814], ..., [ 50. , 84.87456 , 110.11638 ], [ 50. , 110.49849 , 26.430277]], ..., [[ 27.262814, 27.262814, 27.262814], [ 27.262814, 27.262814, 27.262814], ..., [165.49283 , 123.84373 , 50. ], [ 50. , 124.17399 , 50. ]], [[ 27.262814, 27.262814, 27.262814], [ 27.262814, 27.262814, 27.262814], ..., [171.94188 , 44.177135, 50. ], [ 50. , 73.79445 , 50. ]]], dtype=float32) - dgnd_a01_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- m
- long_name :
- dry dgnum, interstitial, mode 01
- basename :
- dgnd_a01
array([[[5.350000e-08, 5.350000e-08, 5.350000e-08], [5.350000e-08, 5.350000e-08, 5.350000e-08], ..., [1.538007e-07, 1.408233e-07, 1.425987e-07], [1.538352e-07, 1.408427e-07, 1.426624e-07]], [[5.350000e-08, 5.350000e-08, 5.350000e-08], [5.350000e-08, 5.350000e-08, 5.350000e-08], ..., [1.535787e-07, 1.404593e-07, 1.428532e-07], [1.536173e-07, 1.404778e-07, 1.429291e-07]], ..., [[5.350000e-08, 5.350000e-08, 5.350000e-08], [5.350000e-08, 5.350000e-08, 5.350000e-08], ..., [1.465580e-07, 1.470669e-07, 1.461231e-07], [1.465735e-07, 1.470895e-07, 1.461770e-07]], [[5.350000e-08, 5.350000e-08, 5.350000e-08], [5.350000e-08, 5.350000e-08, 5.350000e-08], ..., [1.462663e-07, 1.478113e-07, 1.455344e-07], [1.462815e-07, 1.478350e-07, 1.456326e-07]]], dtype=float32) - dgnd_a02_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- m
- long_name :
- dry dgnum, interstitial, mode 02
- basename :
- dgnd_a02
array([[[1.480487e-08, 1.466583e-08, 1.462628e-08], [1.490486e-08, 1.477309e-08, 1.478014e-08], ..., [5.078344e-08, 5.066528e-08, 4.933421e-08], [5.078599e-08, 5.063943e-08, 4.925782e-08]], [[1.481450e-08, 1.475164e-08, 1.471308e-08], [1.500007e-08, 1.486635e-08, 1.487677e-08], ..., [5.089784e-08, 5.046972e-08, 4.907581e-08], [5.090602e-08, 5.044186e-08, 4.897077e-08]], ..., [[1.430434e-08, 1.431591e-08, 1.442154e-08], [1.468673e-08, 1.469754e-08, 1.478303e-08], ..., [4.127254e-08, 5.149756e-08, 5.097303e-08], [4.127606e-08, 5.149648e-08, 5.100145e-08]], [[1.415095e-08, 1.421372e-08, 1.432900e-08], [1.450556e-08, 1.458697e-08, 1.470383e-08], ..., [4.146125e-08, 5.136985e-08, 4.965834e-08], [4.146276e-08, 5.137281e-08, 4.972674e-08]]], dtype=float32) - dgnd_a03_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- m
- long_name :
- dry dgnum, interstitial, mode 03
- basename :
- dgnd_a03
array([[[1.000000e-06, 1.000000e-06, 1.000000e-06], [1.000000e-06, 1.000000e-06, 1.000000e-06], ..., [1.573956e-06, 1.741924e-06, 1.718382e-06], [1.574899e-06, 1.742353e-06, 1.718453e-06]], [[1.000000e-06, 1.000000e-06, 1.000000e-06], [1.000000e-06, 1.000000e-06, 1.000000e-06], ..., [1.557045e-06, 1.764432e-06, 1.735878e-06], [1.557939e-06, 1.765004e-06, 1.735149e-06]], ..., [[1.000000e-06, 1.000000e-06, 1.000000e-06], [1.000000e-06, 1.000000e-06, 1.000000e-06], ..., [1.458210e-06, 1.084148e-06, 1.361927e-06], [1.458812e-06, 1.083809e-06, 1.361246e-06]], [[1.000000e-06, 1.000000e-06, 1.000000e-06], [1.000000e-06, 1.000000e-06, 1.000000e-06], ..., [1.479743e-06, 1.081325e-06, 1.331967e-06], [1.480405e-06, 1.081201e-06, 1.331735e-06]]], dtype=float32) - dgnd_a04_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- m
- long_name :
- dry dgnum, interstitial, mode 04
- basename :
- dgnd_a04
array([[[2.697441e-08, 2.685571e-08, 2.681117e-08], [2.722151e-08, 2.702379e-08, 2.703267e-08], ..., [1.000000e-07, 1.000000e-07, 1.000000e-07], [1.000000e-07, 1.000000e-07, 1.000000e-07]], [[2.698767e-08, 2.690567e-08, 2.688100e-08], [2.735804e-08, 2.712619e-08, 2.715915e-08], ..., [1.000000e-07, 1.000000e-07, 1.000000e-07], [1.000000e-07, 1.000000e-07, 1.000000e-07]], ..., [[2.614744e-08, 2.618256e-08, 2.637763e-08], [2.685920e-08, 2.690179e-08, 2.708947e-08], ..., [1.000000e-07, 1.000000e-07, 1.000000e-07], [1.000000e-07, 1.000000e-07, 1.000000e-07]], [[2.585942e-08, 2.597032e-08, 2.619201e-08], [2.650274e-08, 2.669448e-08, 2.691732e-08], ..., [1.000000e-07, 1.000000e-07, 1.000000e-07], [1.000000e-07, 1.000000e-07, 1.000000e-07]]], dtype=float32) - dgnw_a01_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- m
- long_name :
- wet dgnum, interstitial, mode 01
- basename :
- dgnw_a01
array([[[5.350000e-08, 5.350000e-08, 5.350000e-08], [5.350000e-08, 5.350000e-08, 5.350000e-08], ..., [2.251155e-07, 1.831989e-07, 2.026375e-07], [2.198936e-07, 1.796984e-07, 2.007037e-07]], [[5.350000e-08, 5.350000e-08, 5.350000e-08], [5.350000e-08, 5.350000e-08, 5.350000e-08], ..., [2.217218e-07, 1.862384e-07, 2.053802e-07], [2.169189e-07, 1.823520e-07, 2.032737e-07]], ..., [[5.350000e-08, 5.350000e-08, 5.350000e-08], [5.350000e-08, 5.350000e-08, 5.350000e-08], ..., [2.461741e-07, 2.396030e-07, 2.299362e-07], [2.379702e-07, 2.297223e-07, 2.218023e-07]], [[5.350000e-08, 5.350000e-08, 5.350000e-08], [5.350000e-08, 5.350000e-08, 5.350000e-08], ..., [2.479562e-07, 2.491068e-07, 2.383307e-07], [2.395002e-07, 2.380186e-07, 2.281840e-07]]], dtype=float32) - dgnw_a02_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- m
- long_name :
- wet dgnum, interstitial, mode 02
- basename :
- dgnw_a02
array([[[1.480487e-08, 1.466583e-08, 1.462628e-08], [1.490486e-08, 1.477309e-08, 1.478014e-08], ..., [7.532335e-08, 6.683381e-08, 7.115688e-08], [7.364291e-08, 6.550090e-08, 7.036680e-08]], [[1.481450e-08, 1.475164e-08, 1.471308e-08], [1.500007e-08, 1.486635e-08, 1.487677e-08], ..., [7.453830e-08, 6.787982e-08, 7.169922e-08], [7.298254e-08, 6.640958e-08, 7.080743e-08]], ..., [[1.430434e-08, 1.431591e-08, 1.442154e-08], [1.468673e-08, 1.469754e-08, 1.478303e-08], ..., [6.772799e-08, 8.303051e-08, 7.993049e-08], [6.570502e-08, 7.988337e-08, 7.737666e-08]], [[1.415095e-08, 1.421372e-08, 1.432900e-08], [1.450556e-08, 1.458697e-08, 1.470383e-08], ..., [6.817191e-08, 8.590186e-08, 8.066085e-08], [6.609820e-08, 8.238950e-08, 7.763566e-08]]], dtype=float32) - dgnw_a03_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- m
- long_name :
- wet dgnum, interstitial, mode 03
- basename :
- dgnw_a03
array([[[1.000000e-06, 1.000000e-06, 1.000000e-06], [1.000000e-06, 1.000000e-06, 1.000000e-06], ..., [3.077505e-06, 2.855773e-06, 3.213560e-06], [2.984321e-06, 2.776578e-06, 3.170050e-06]], [[1.000000e-06, 1.000000e-06, 1.000000e-06], [1.000000e-06, 1.000000e-06, 1.000000e-06], ..., [3.004940e-06, 2.958761e-06, 3.290406e-06], [2.918796e-06, 2.871633e-06, 3.241364e-06]], ..., [[1.000000e-06, 1.000000e-06, 1.000000e-06], [1.000000e-06, 1.000000e-06, 1.000000e-06], ..., [3.043695e-06, 2.385098e-06, 3.023633e-06], [2.926157e-06, 2.262859e-06, 2.883197e-06]], [[1.000000e-06, 1.000000e-06, 1.000000e-06], [1.000000e-06, 1.000000e-06, 1.000000e-06], ..., [3.100269e-06, 2.432380e-06, 3.068967e-06], [2.978587e-06, 2.301452e-06, 2.902260e-06]]], dtype=float32) - dgnw_a04_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- m
- long_name :
- wet dgnum, interstitial, mode 04
- basename :
- dgnw_a04
array([[[2.697441e-08, 2.685571e-08, 2.681117e-08], [2.722151e-08, 2.702379e-08, 2.703267e-08], ..., [1.000002e-07, 1.000001e-07, 1.000000e-07], [1.000002e-07, 1.000000e-07, 1.000000e-07]], [[2.698767e-08, 2.690567e-08, 2.688100e-08], [2.735804e-08, 2.712619e-08, 2.715915e-08], ..., [1.000002e-07, 1.000001e-07, 1.000000e-07], [1.000002e-07, 1.000001e-07, 1.000000e-07]], ..., [[2.614744e-08, 2.618256e-08, 2.637763e-08], [2.685920e-08, 2.690179e-08, 2.708947e-08], ..., [1.000011e-07, 1.000003e-07, 1.000002e-07], [1.000010e-07, 1.000002e-07, 1.000002e-07]], [[2.585942e-08, 2.597032e-08, 2.619201e-08], [2.650274e-08, 2.669448e-08, 2.691732e-08], ..., [1.000012e-07, 1.000003e-07, 1.000002e-07], [1.000010e-07, 1.000002e-07, 1.000002e-07]]], dtype=float32) - dst_a1_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- dst_a1 concentration
- basename :
- dst_a1
array([[[5.804864e-21, 7.652429e-21, 7.801187e-21], [8.460199e-21, 7.884959e-21, 7.162307e-21], ..., [4.232259e-12, 6.476996e-12, 8.375067e-12], [4.227744e-12, 6.471166e-12, 8.378413e-12]], [[5.933393e-21, 5.452369e-21, 5.834617e-21], [7.812916e-21, 5.862707e-21, 5.542979e-21], ..., [4.227209e-12, 5.922610e-12, 7.934459e-12], [4.224422e-12, 5.917231e-12, 7.934624e-12]], ..., [[2.305210e-21, 2.017200e-21, 2.112161e-21], [4.905886e-21, 4.683739e-21, 5.167591e-21], ..., [2.547932e-12, 9.768533e-12, 1.033967e-11], [2.549328e-12, 9.770090e-12, 1.035785e-11]], [[2.086260e-21, 1.981785e-21, 2.129155e-21], [3.923488e-21, 4.190608e-21, 4.621605e-21], ..., [2.211296e-12, 8.291134e-12, 8.624711e-12], [2.212167e-12, 8.300520e-12, 8.659100e-12]]], dtype=float32) - dst_a3_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- dst_a3 concentration
- basename :
- dst_a3
array([[[1.000000e-36, 1.008142e-36, 1.017086e-36], [1.000506e-36, 1.000000e-36, 1.000000e-36], ..., [9.111863e-12, 3.349119e-11, 4.796853e-11], [9.090425e-12, 3.348439e-11, 4.767653e-11]], [[1.000000e-36, 1.006386e-36, 1.023065e-36], [1.000197e-36, 1.000000e-36, 1.000000e-36], ..., [8.646793e-12, 3.051792e-11, 4.501007e-11], [8.637558e-12, 3.052418e-11, 4.461893e-11]], ..., [[1.002299e-36, 1.004569e-36, 1.008369e-36], [1.000143e-36, 1.000065e-36, 1.000000e-36], ..., [9.353129e-12, 4.287472e-11, 5.072404e-11], [9.325236e-12, 4.288529e-11, 5.082225e-11]], [[1.005624e-36, 1.001142e-36, 1.004832e-36], [1.000000e-36, 1.000637e-36, 1.000000e-36], ..., [9.513604e-12, 3.444415e-11, 4.027499e-11], [9.465859e-12, 3.449150e-11, 4.053228e-11]]], dtype=float32) - dst_c1_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- dst_c1 in cloud water
- basename :
- dst_c1
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - dst_c3_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- dst_c3 in cloud water
- basename :
- dst_c3
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - lambda_cloud_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- 1/meter
- long_name :
- lambda in cloud
- basename :
- lambda_cloud
array([[[ 0., 0., 0.], [ 0., 0., 0.], ..., [252000., 0., 0.], [252000., 0., 0.]], [[ 0., 0., 0.], [ 0., 0., 0.], ..., [252000., 0., 0.], [252000., 0., 0.]], ..., [[ 0., 0., 0.], [ 0., 0., 0.], ..., [ 0., 0., 252000.], [252000., 0., 252000.]], [[ 0., 0., 0.], [ 0., 0., 0.], ..., [ 0., 0., 252000.], [252000., 0., 252000.]]], dtype=float32) - mom_a1_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- mom_a1 concentration
- basename :
- mom_a1
array([[[4.131965e-26, 6.302358e-26, 6.575023e-26], [6.189963e-26, 6.104222e-26, 5.330224e-26], ..., [6.720337e-12, 8.400120e-12, 1.044640e-11], [6.739742e-12, 8.413766e-12, 1.044513e-11]], [[4.188877e-26, 4.166556e-26, 4.701396e-26], [5.516865e-26, 4.092375e-26, 3.757598e-26], ..., [6.214099e-12, 8.027821e-12, 1.009871e-11], [6.230851e-12, 8.040790e-12, 1.009322e-11]], ..., [[1.342985e-26, 1.116637e-26, 1.163141e-26], [3.113532e-26, 2.985537e-26, 3.327954e-26], ..., [1.193179e-10, 7.832813e-11, 3.609081e-11], [1.196749e-10, 7.839725e-11, 3.614348e-11]], [[1.241006e-26, 1.127049e-26, 1.201095e-26], [2.441700e-26, 2.643050e-26, 2.975126e-26], ..., [1.095913e-10, 8.218459e-11, 3.656975e-11], [1.099319e-10, 8.229418e-11, 3.668588e-11]]], dtype=float32) - mom_a2_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- mom_a2 concentration
- basename :
- mom_a2
array([[[1.010062e-20, 9.711989e-21, 9.673306e-21], [1.049559e-20, 9.944220e-21, 1.000503e-20], ..., [3.828882e-14, 4.752484e-14, 5.781049e-14], [3.869569e-14, 4.779978e-14, 5.764908e-14]], [[1.015522e-20, 1.017355e-20, 1.019305e-20], [1.093655e-20, 1.040312e-20, 1.050547e-20], ..., [3.039366e-14, 4.145218e-14, 5.194101e-14], [3.072369e-14, 4.170965e-14, 5.175515e-14]], ..., [[8.001083e-21, 8.148076e-21, 8.672608e-21], [9.443167e-21, 9.515345e-21, 9.861222e-21], ..., [7.727504e-13, 4.026364e-13, 1.748360e-13], [7.785473e-13, 4.040722e-13, 1.750037e-13]], [[7.339939e-21, 7.592763e-21, 8.160481e-21], [8.591991e-21, 8.976936e-21, 9.519785e-21], ..., [6.731958e-13, 3.587924e-13, 1.341744e-13], [6.785310e-13, 3.603460e-13, 1.351618e-13]]], dtype=float32) - mom_a3_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- mom_a3 concentration
- basename :
- mom_a3
array([[[1.000000e-36, 1.008142e-36, 1.017086e-36], [1.000506e-36, 1.000000e-36, 1.000000e-36], ..., [2.585518e-13, 1.286249e-12, 1.979052e-12], [2.536274e-13, 1.290075e-12, 1.942979e-12]], [[1.000000e-36, 1.006386e-36, 1.023065e-36], [1.000197e-36, 1.000000e-36, 1.000000e-36], ..., [2.512782e-13, 1.527970e-12, 2.166763e-12], [2.478317e-13, 1.534234e-12, 2.113664e-12]], ..., [[1.002299e-36, 1.004569e-36, 1.008369e-36], [1.000143e-36, 1.000065e-36, 1.000000e-36], ..., [6.349254e-13, 6.009559e-13, 6.054745e-13], [6.298187e-13, 5.959219e-13, 6.080172e-13]], [[1.005624e-36, 1.001142e-36, 1.004832e-36], [1.000000e-36, 1.000637e-36, 1.000000e-36], ..., [6.824809e-13, 5.453982e-13, 5.080338e-13], [6.754721e-13, 5.452080e-13, 5.133212e-13]]], dtype=float32) - mom_a4_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- mom_a4 concentration
- basename :
- mom_a4
array([[[4.006153e-23, 3.842548e-23, 3.825403e-23], [4.195545e-23, 3.949100e-23, 3.970387e-23], ..., [5.883083e-17, 6.053436e-17, 4.848622e-17], [5.876967e-17, 6.052565e-17, 4.843298e-17]], [[4.034593e-23, 4.016161e-23, 4.021369e-23], [4.383004e-23, 4.131424e-23, 4.173413e-23], ..., [5.925158e-17, 6.399712e-17, 5.252741e-17], [5.919070e-17, 6.398348e-17, 5.244511e-17]], ..., [[3.165183e-23, 3.219448e-23, 3.425849e-23], [3.781796e-23, 3.811410e-23, 3.972177e-23], ..., [1.616892e-16, 1.317090e-16, 1.161282e-16], [1.616805e-16, 1.316720e-16, 1.159665e-16]], [[2.905350e-23, 3.007716e-23, 3.232084e-23], [3.431524e-23, 3.597667e-23, 3.824513e-23], ..., [1.928930e-16, 1.305467e-16, 1.215699e-16], [1.928643e-16, 1.304623e-16, 1.213834e-16]]], dtype=float32) - mom_c1_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- mom_c1 in cloud water
- basename :
- mom_c1
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - mom_c2_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- mom_c2 in cloud water
- basename :
- mom_c2
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - mom_c3_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- mom_c3 in cloud water
- basename :
- mom_c3
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - mom_c4_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- mom_c4 in cloud water
- basename :
- mom_c4
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - mu_cloud_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- 1
- long_name :
- mu in cloud
- basename :
- mu_cloud
array([[[0. , 0. , 0. ], [0. , 0. , 0. ], ..., [5.3, 0. , 0. ], [5.3, 0. , 0. ]], [[0. , 0. , 0. ], [0. , 0. , 0. ], ..., [5.3, 0. , 0. ], [5.3, 0. , 0. ]], ..., [[0. , 0. , 0. ], [0. , 0. , 0. ], ..., [0. , 0. , 5.3], [5.3, 0. , 5.3]], [[0. , 0. , 0. ], [0. , 0. , 0. ], ..., [0. , 0. , 5.3], [5.3, 0. , 5.3]]], dtype=float32) - ncl_a1_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- ncl_a1 concentration
- basename :
- ncl_a1
array([[[3.704289e-21, 5.547556e-21, 5.732633e-21], [5.692622e-21, 5.541356e-21, 4.858783e-21], ..., [4.312804e-11, 5.225967e-11, 5.861165e-11], [4.320103e-11, 5.233718e-11, 5.880093e-11]], [[3.843606e-21, 3.617797e-21, 4.041520e-21], [5.021757e-21, 3.618545e-21, 3.319048e-21], ..., [4.047085e-11, 4.984666e-11, 5.630644e-11], [4.053752e-11, 4.992371e-11, 5.649558e-11]], ..., [[1.305000e-21, 1.076865e-21, 1.105782e-21], [3.051720e-21, 2.959224e-21, 3.268604e-21], ..., [1.553401e-10, 1.052198e-10, 5.746668e-11], [1.559314e-10, 1.054895e-10, 5.776408e-11]], [[1.220382e-21, 1.098210e-21, 1.154636e-21], [2.417424e-21, 2.622880e-21, 2.909019e-21], ..., [1.486926e-10, 1.121994e-10, 5.404013e-11], [1.492769e-10, 1.125308e-10, 5.444842e-11]]], dtype=float32) - ncl_a2_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- ncl_a2 concentration
- basename :
- ncl_a2
array([[[5.295140e-16, 5.056154e-16, 5.029282e-16], [5.467271e-16, 5.180335e-16, 5.209739e-16], ..., [1.835175e-13, 2.931254e-13, 3.500736e-13], [1.850207e-13, 2.948350e-13, 3.523211e-13]], [[5.316725e-16, 5.329235e-16, 5.326304e-16], [5.694357e-16, 5.434870e-16, 5.485416e-16], ..., [1.487721e-13, 2.606421e-13, 3.180104e-13], [1.499663e-13, 2.623067e-13, 3.205303e-13]], ..., [[4.214250e-16, 4.288181e-16, 4.553402e-16], [4.929167e-16, 4.958011e-16, 5.105920e-16], ..., [1.361883e-12, 6.738093e-13, 3.202849e-13], [1.373268e-12, 6.778302e-13, 3.227321e-13]], [[3.874486e-16, 4.011474e-16, 4.300488e-16], [4.511987e-16, 4.691859e-16, 4.952863e-16], ..., [1.277437e-12, 6.696059e-13, 2.716707e-13], [1.288601e-12, 6.741594e-13, 2.751628e-13]]], dtype=float32) - ncl_a3_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- ncl_a3 concentration
- basename :
- ncl_a3
array([[[1.000000e-36, 1.008142e-36, 1.017086e-36], [1.000506e-36, 1.000000e-36, 1.000000e-36], ..., [8.350409e-10, 1.427077e-09, 1.587818e-09], [8.369200e-10, 1.430396e-09, 1.590910e-09]], [[1.000000e-36, 1.006386e-36, 1.023065e-36], [1.000197e-36, 1.000000e-36, 1.000000e-36], ..., [7.532723e-10, 1.399280e-09, 1.562210e-09], [7.549908e-10, 1.402916e-09, 1.563235e-09]], ..., [[1.002299e-36, 1.004569e-36, 1.008369e-36], [1.000143e-36, 1.000065e-36, 1.000000e-36], ..., [2.537509e-09, 7.163339e-10, 7.730431e-10], [2.551266e-09, 7.178408e-10, 7.763071e-10]], [[1.005624e-36, 1.001142e-36, 1.004832e-36], [1.000000e-36, 1.000637e-36, 1.000000e-36], ..., [2.511978e-09, 7.591464e-10, 6.735736e-10], [2.526009e-09, 7.615767e-10, 6.789608e-10]]], dtype=float32) - ncl_c1_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- ncl_c1 in cloud water
- basename :
- ncl_c1
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - ncl_c2_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- ncl_c2 in cloud water
- basename :
- ncl_c2
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - ncl_c3_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- ncl_c3 in cloud water
- basename :
- ncl_c3
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - num_a1_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- 1/kg
- long_name :
- num_a1 concentration
- basename :
- num_a1
array([[[1.338253e-01, 1.717254e-01, 1.772148e-01], [1.957873e-01, 1.676273e-01, 1.497141e-01], ..., [1.861399e+07, 2.060367e+07, 2.314722e+07], [1.859918e+07, 2.058833e+07, 2.313810e+07]], [[1.417414e-01, 1.206110e-01, 1.285304e-01], [1.876392e-01, 1.331366e-01, 1.255331e-01], ..., [1.875360e+07, 1.980489e+07, 2.243906e+07], [1.874246e+07, 1.978892e+07, 2.241760e+07]], ..., [[5.076634e-02, 4.333969e-02, 4.450741e-02], [1.290688e-01, 1.224868e-01, 1.294378e-01], ..., [3.023046e+07, 3.557735e+07, 3.073835e+07], [3.028959e+07, 3.558006e+07, 3.078262e+07]], [[4.726077e-02, 4.370632e-02, 4.611306e-02], [1.124416e-01, 1.126632e-01, 1.168149e-01], ..., [2.840927e+07, 3.458664e+07, 2.805029e+07], [2.846806e+07, 3.461170e+07, 2.812799e+07]]], dtype=float32) - num_a2_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- 1/kg
- long_name :
- num_a2 concentration
- basename :
- num_a2
array([[[ 4295866. , 4169037.2, 4173743. ], [ 4350748.5, 4200435.5, 4219150.5], ..., [10027550. , 15829427. , 22187108. ], [10024589. , 15859493. , 22432274. ]], [[ 4314747.5, 4340131. , 4358134. ], [ 4461686. , 4345496. , 4376109. ], ..., [10030834. , 15593760. , 22343884. ], [10026496. , 15626587. , 22667688. ]], ..., [[ 3774417.2, 3823092.2, 3953772. ], [ 4077168.5, 4086484.8, 4134859.5], ..., [32919884. , 22649126. , 22522964. ], [32994956. , 22667226. , 22555044. ]], [[ 3605583.5, 3677227.5, 3829992.8], [ 3888963.5, 3971150.2, 4083001.2], ..., [31634676. , 22328604. , 20844644. ], [31712162. , 22356636. , 20903662. ]]], dtype=float32) - num_a3_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- 1/kg
- long_name :
- num_a3 concentration
- basename :
- num_a3
array([[[1.000000e-05, 1.000000e-05, 1.000000e-05], [1.000000e-05, 1.000000e-05, 1.000000e-05], ..., [4.628726e+04, 5.986064e+04, 6.938585e+04], [4.635316e+04, 5.995389e+04, 6.967086e+04]], [[1.000000e-05, 1.000000e-05, 1.000000e-05], [1.000000e-05, 1.000000e-05, 1.000000e-05], ..., [4.328376e+04, 5.676087e+04, 6.636424e+04], [4.334311e+04, 5.685349e+04, 6.665691e+04]], ..., [[1.000000e-05, 1.000000e-05, 1.000000e-05], [1.000000e-05, 1.000000e-05, 1.000000e-05], ..., [1.760289e+05, 1.272449e+05, 6.956682e+04], [1.767665e+05, 1.276186e+05, 6.995831e+04]], [[1.000000e-05, 1.000000e-05, 1.000000e-05], [1.000000e-05, 1.000000e-05, 1.000000e-05], ..., [1.666510e+05, 1.343450e+05, 6.454646e+04], [1.673641e+05, 1.347976e+05, 6.509211e+04]]], dtype=float32) - num_a4_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- 1/kg
- long_name :
- num_a4 concentration
- basename :
- num_a4
array([[[1.220546e+03, 1.183775e+03, 1.184235e+03], [1.244847e+03, 1.195431e+03, 1.200330e+03], ..., [2.312570e+06, 4.824026e+06, 7.515636e+06], [2.310081e+06, 4.838560e+06, 7.673856e+06]], [[1.228688e+03, 1.230159e+03, 1.234260e+03], [1.281392e+03, 1.236980e+03, 1.244958e+03], ..., [2.402354e+06, 5.194236e+06, 8.162666e+06], [2.400098e+06, 5.209910e+06, 8.367775e+06]], ..., [[1.061129e+03, 1.073977e+03, 1.115421e+03], [1.168437e+03, 1.171920e+03, 1.196439e+03], ..., [1.054849e+06, 4.333652e+06, 6.568827e+06], [1.054844e+06, 4.344556e+06, 6.608746e+06]], [[1.009126e+03, 1.030727e+03, 1.077700e+03], [1.106368e+03, 1.134999e+03, 1.174893e+03], ..., [1.285064e+06, 4.081618e+06, 5.547981e+06], [1.284749e+06, 4.092146e+06, 5.598782e+06]]], dtype=float32) - num_c1_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- 1/kg
- long_name :
- num_c1 in cloud water
- basename :
- num_c1
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - num_c2_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- 1/kg
- long_name :
- num_c2 in cloud water
- basename :
- num_c2
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - num_c3_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- 1/kg
- long_name :
- num_c3 in cloud water
- basename :
- num_c3
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - num_c4_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- 1/kg
- long_name :
- num_c4 in cloud water
- basename :
- num_c4
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - pom_a1_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- pom_a1 concentration
- basename :
- pom_a1
array([[[6.421508e-21, 8.547237e-21, 8.737880e-21], [9.809280e-21, 8.913517e-21, 8.044026e-21], ..., [2.445058e-12, 4.338630e-12, 5.527486e-12], [2.438154e-12, 4.333989e-12, 5.531359e-12]], [[6.738655e-21, 5.881500e-21, 6.296602e-21], [8.879888e-21, 6.409733e-21, 6.014598e-21], ..., [2.443407e-12, 3.960072e-12, 5.189731e-12], [2.436743e-12, 3.955290e-12, 5.191609e-12]], ..., [[2.722642e-21, 2.349295e-21, 2.417572e-21], [5.908266e-21, 5.738134e-21, 6.260715e-21], ..., [1.868171e-12, 7.433071e-12, 8.224960e-12], [1.868973e-12, 7.434636e-12, 8.242127e-12]], [[2.498192e-21, 2.345076e-21, 2.489984e-21], [4.783662e-21, 5.140967e-21, 5.586217e-21], ..., [1.593162e-12, 6.329831e-12, 6.858975e-12], [1.593474e-12, 6.337117e-12, 6.890305e-12]]], dtype=float32) - pom_a3_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- pom_a3 concentration
- basename :
- pom_a3
array([[[1.000000e-36, 1.008142e-36, 1.017086e-36], [1.000506e-36, 1.000000e-36, 1.000000e-36], ..., [1.119811e-13, 7.706562e-13, 1.217095e-12], [1.100097e-13, 7.726260e-13, 1.176527e-12]], [[1.000000e-36, 1.006386e-36, 1.023065e-36], [1.000197e-36, 1.000000e-36, 1.000000e-36], ..., [1.035906e-13, 8.975394e-13, 1.314231e-12], [1.020221e-13, 9.009420e-13, 1.262572e-12]], ..., [[1.002299e-36, 1.004569e-36, 1.008369e-36], [1.000143e-36, 1.000065e-36, 1.000000e-36], ..., [1.246989e-13, 2.328651e-13, 2.756253e-13], [1.235947e-13, 2.320225e-13, 2.765762e-13]], [[1.005624e-36, 1.001142e-36, 1.004832e-36], [1.000000e-36, 1.000637e-36, 1.000000e-36], ..., [1.589688e-13, 1.851450e-13, 2.105609e-13], [1.569745e-13, 1.852339e-13, 2.126044e-13]]], dtype=float32) - pom_a4_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- pom_a4 concentration
- basename :
- pom_a4
array([[[3.292528e-17, 3.150814e-17, 3.136328e-17], [3.451287e-17, 3.242213e-17, 3.258661e-17], ..., [2.184692e-12, 4.198049e-12, 6.363732e-12], [2.182420e-12, 4.210124e-12, 6.496192e-12]], [[3.319606e-17, 3.292663e-17, 3.294372e-17], [3.606407e-17, 3.393359e-17, 3.427703e-17], ..., [2.278241e-12, 4.560370e-12, 6.895161e-12], [2.276164e-12, 4.573301e-12, 7.065205e-12]], ..., [[2.607521e-17, 2.649581e-17, 2.813409e-17], [3.111743e-17, 3.135905e-17, 3.269037e-17], ..., [1.503935e-12, 4.548177e-12, 6.249700e-12], [1.504096e-12, 4.560083e-12, 6.288756e-12]], [[2.399026e-17, 2.481927e-17, 2.661725e-17], [2.831246e-17, 2.968008e-17, 3.149530e-17], ..., [1.757661e-12, 4.394444e-12, 5.413277e-12], [1.757482e-12, 4.406107e-12, 5.462001e-12]]], dtype=float32) - pom_c1_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- pom_c1 in cloud water
- basename :
- pom_c1
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - pom_c3_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- pom_c3 in cloud water
- basename :
- pom_c3
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - pom_c4_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- pom_c4 in cloud water
- basename :
- pom_c4
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - so4_a1_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- so4_a1 concentration
- basename :
- so4_a1
array([[[1.775279e-20, 2.675850e-20, 2.762384e-20], [2.770446e-20, 2.697202e-20, 2.365226e-20], ..., [9.184924e-11, 5.130159e-11, 5.233505e-11], [9.182483e-11, 5.124646e-11, 5.234477e-11]], [[1.857201e-20, 1.725982e-20, 1.930478e-20], [2.422490e-20, 1.728413e-20, 1.581224e-20], ..., [9.361488e-11, 5.228755e-11, 5.420090e-11], [9.361124e-11, 5.222418e-11, 5.417721e-11]], ..., [[6.415439e-21, 5.280673e-21, 5.401717e-21], [1.506247e-20, 1.473030e-20, 1.626745e-20], ..., [8.360907e-11, 9.214895e-11, 8.767217e-11], [8.365871e-11, 9.208014e-11, 8.794991e-11]], [[6.021859e-21, 5.409724e-21, 5.673026e-21], [1.191411e-20, 1.303513e-20, 1.446577e-20], ..., [8.145994e-11, 1.073031e-10, 8.923800e-11], [8.152470e-11, 1.072962e-10, 8.969181e-11]]], dtype=float32) - so4_a2_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- so4_a2 concentration
- basename :
- so4_a2
array([[[3.394394e-14, 3.205548e-14, 3.183891e-14], [3.507727e-14, 3.299540e-14, 3.319369e-14], ..., [2.247730e-12, 3.719043e-12, 4.648935e-12], [2.248924e-12, 3.721015e-12, 4.690700e-12]], [[3.415670e-14, 3.395105e-14, 3.383550e-14], [3.665999e-14, 3.478266e-14, 3.510290e-14], ..., [2.298861e-12, 3.775903e-12, 4.775734e-12], [2.299966e-12, 3.778858e-12, 4.827933e-12]], ..., [[2.693570e-14, 2.735425e-14, 2.892978e-14], [3.148348e-14, 3.162746e-14, 3.256847e-14], ..., [3.338851e-12, 4.388640e-12, 3.948487e-12], [3.343773e-12, 4.387807e-12, 3.968799e-12]], [[2.490104e-14, 2.574172e-14, 2.747761e-14], [2.892643e-14, 3.004255e-14, 3.164704e-14], ..., [3.325998e-12, 5.036674e-12, 3.654928e-12], [3.331345e-12, 5.040690e-12, 3.687201e-12]]], dtype=float32) - so4_a3_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- so4_a3 concentration
- basename :
- so4_a3
array([[[8.919359e-32, 8.713277e-32, 8.859738e-32], [1.194916e-31, 1.039095e-31, 9.259755e-32], ..., [5.036401e-12, 8.045981e-12, 1.049921e-11], [4.967601e-12, 8.071658e-12, 1.021057e-11]], [[8.495710e-32, 8.879492e-32, 9.092623e-32], [1.109899e-31, 1.030225e-31, 8.804732e-32], ..., [4.960293e-12, 1.041905e-11, 1.250750e-11], [4.905678e-12, 1.046561e-11, 1.206574e-11]], ..., [[8.460622e-32, 8.912467e-32, 8.800870e-32], [7.326924e-32, 6.320539e-32, 6.562081e-32], ..., [1.733168e-12, 1.996243e-12, 2.321212e-12], [1.715461e-12, 1.984839e-12, 2.331160e-12]], [[8.196245e-32, 8.916084e-32, 8.885043e-32], [6.436008e-32, 7.546833e-32, 7.640161e-32], ..., [2.114112e-12, 1.720037e-12, 1.829379e-12], [2.084043e-12, 1.720043e-12, 1.849771e-12]]], dtype=float32) - so4_c1_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- so4_c1 in cloud water
- basename :
- so4_c1
array([[[3.973878e-20, 3.973878e-20, 3.973878e-20], [3.973878e-20, 3.973878e-20, 3.973878e-20], ..., [3.974574e-20, 3.974157e-20, 3.973883e-20], [3.975725e-20, 3.974157e-20, 3.973918e-20]], [[3.973878e-20, 3.973878e-20, 3.973878e-20], [3.973878e-20, 3.973878e-20, 3.973878e-20], ..., [3.974556e-20, 3.974164e-20, 3.973882e-20], [3.975794e-20, 3.974164e-20, 3.973911e-20]], ..., [[3.973878e-20, 3.973878e-20, 3.973878e-20], [3.973878e-20, 3.973878e-20, 3.973878e-20], ..., [3.974047e-20, 3.973907e-20, 3.973878e-20], [3.974250e-20, 3.973940e-20, 3.973878e-20]], [[3.973878e-20, 3.973878e-20, 3.973878e-20], [3.973878e-20, 3.973878e-20, 3.973878e-20], ..., [3.974054e-20, 3.973953e-20, 3.973878e-20], [3.974271e-20, 3.974041e-20, 3.973878e-20]]], dtype=float32) - so4_c2_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- so4_c2 in cloud water
- basename :
- so4_c2
array([[[3.973878e-20, 3.973878e-20, 3.973878e-20], [3.973878e-20, 3.973878e-20, 3.973878e-20], ..., [3.973181e-20, 3.973598e-20, 3.973872e-20], [3.972031e-20, 3.973598e-20, 3.973837e-20]], [[3.973878e-20, 3.973878e-20, 3.973878e-20], [3.973878e-20, 3.973878e-20, 3.973878e-20], ..., [3.973199e-20, 3.973591e-20, 3.973874e-20], [3.971962e-20, 3.973591e-20, 3.973844e-20]], ..., [[3.973878e-20, 3.973878e-20, 3.973878e-20], [3.973878e-20, 3.973878e-20, 3.973878e-20], ..., [3.973708e-20, 3.973848e-20, 3.973878e-20], [3.973506e-20, 3.973815e-20, 3.973878e-20]], [[3.973878e-20, 3.973878e-20, 3.973878e-20], [3.973878e-20, 3.973878e-20, 3.973878e-20], ..., [3.973702e-20, 3.973802e-20, 3.973878e-20], [3.973485e-20, 3.973715e-20, 3.973878e-20]]], dtype=float32) - so4_c3_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- so4_c3 in cloud water
- basename :
- so4_c3
array([[[3.973878e-20, 3.973878e-20, 3.973878e-20], [3.973878e-20, 3.973878e-20, 3.973878e-20], ..., [3.973878e-20, 3.973878e-20, 3.973878e-20], [3.973878e-20, 3.973878e-20, 3.973878e-20]], [[3.973878e-20, 3.973878e-20, 3.973878e-20], [3.973878e-20, 3.973878e-20, 3.973878e-20], ..., [3.973878e-20, 3.973878e-20, 3.973878e-20], [3.973878e-20, 3.973878e-20, 3.973878e-20]], ..., [[3.973878e-20, 3.973878e-20, 3.973878e-20], [3.973878e-20, 3.973878e-20, 3.973878e-20], ..., [3.973878e-20, 3.973878e-20, 3.973878e-20], [3.973878e-20, 3.973878e-20, 3.973878e-20]], [[3.973878e-20, 3.973878e-20, 3.973878e-20], [3.973878e-20, 3.973878e-20, 3.973878e-20], ..., [3.973878e-20, 3.973878e-20, 3.973878e-20], [3.973878e-20, 3.973878e-20, 3.973878e-20]]], dtype=float32) - soa_a1_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- soa_a1 concentration
- basename :
- soa_a1
array([[[1.011831e-21, 1.251514e-21, 1.280989e-21], [1.575085e-21, 1.380106e-21, 1.222940e-21], ..., [8.405968e-11, 7.289953e-11, 8.920702e-11], [8.399879e-11, 7.283855e-11, 8.924628e-11]], [[1.081241e-21, 8.507416e-22, 8.857639e-22], [1.313650e-21, 8.844142e-22, 8.183262e-22], ..., [8.526185e-11, 6.785436e-11, 8.582083e-11], [8.524231e-11, 6.779038e-11, 8.582803e-11]], ..., [[4.721835e-22, 3.744191e-22, 3.608244e-22], [1.139342e-21, 1.145632e-21, 1.223593e-21], ..., [2.943124e-11, 1.115082e-10, 1.213537e-10], [2.938569e-11, 1.114978e-10, 1.215776e-10]], [[4.821487e-22, 4.000795e-22, 3.894274e-22], [9.716924e-22, 1.010242e-21, 1.080195e-21], ..., [2.511042e-11, 9.514413e-11, 1.009620e-10], [2.506064e-11, 9.521405e-11, 1.013850e-10]]], dtype=float32) - soa_a2_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- soa_a2 concentration
- basename :
- soa_a2
array([[[2.672530e-16, 2.302480e-16, 2.247139e-16], [2.773457e-16, 2.471573e-16, 2.464871e-16], ..., [4.781981e-13, 6.378493e-13, 9.365810e-13], [4.754140e-13, 6.360676e-13, 9.355514e-13]], [[2.713140e-16, 2.505061e-16, 2.429230e-16], [2.940838e-16, 2.634995e-16, 2.650592e-16], ..., [4.846218e-13, 5.503163e-13, 8.534588e-13], [4.825514e-13, 5.481655e-13, 8.523132e-13]], ..., [[1.906364e-16, 1.902952e-16, 1.946812e-16], [2.279651e-16, 2.273309e-16, 2.312341e-16], ..., [2.088122e-13, 1.294505e-12, 1.721013e-12], [2.020897e-13, 1.294417e-12, 1.720704e-12]], [[1.836989e-16, 1.858141e-16, 1.922736e-16], [2.138575e-16, 2.190767e-16, 2.244497e-16], ..., [2.390401e-13, 8.650764e-13, 1.329345e-12], [2.322701e-13, 8.646071e-13, 1.333208e-12]]], dtype=float32) - soa_a3_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- soa_a3 concentration
- basename :
- soa_a3
array([[[1.000000e-36, 1.008142e-36, 1.017086e-36], [1.000506e-36, 1.000000e-36, 1.000000e-36], ..., [5.327910e-12, 1.380781e-11, 2.011213e-11], [5.266487e-12, 1.384208e-11, 1.954086e-11]], [[1.000000e-36, 1.006386e-36, 1.023065e-36], [1.000197e-36, 1.000000e-36, 1.000000e-36], ..., [5.217372e-12, 1.615597e-11, 2.215671e-11], [5.170241e-12, 1.621465e-11, 2.139680e-11]], ..., [[1.002299e-36, 1.004569e-36, 1.008369e-36], [1.000143e-36, 1.000065e-36, 1.000000e-36], ..., [1.861219e-12, 4.376605e-12, 5.312397e-12], [1.840191e-12, 4.365021e-12, 5.328454e-12]], [[1.005624e-36, 1.001142e-36, 1.004832e-36], [1.000000e-36, 1.000637e-36, 1.000000e-36], ..., [2.141626e-12, 3.464459e-12, 4.076560e-12], [2.112008e-12, 3.464705e-12, 4.111061e-12]]], dtype=float32) - soa_c1_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- soa_c1 in cloud water
- basename :
- soa_c1
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - soa_c2_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- soa_c2 in cloud water
- basename :
- soa_c2
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - soa_c3_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- kg/kg
- long_name :
- soa_c3 in cloud water
- basename :
- soa_c3
array([[[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], ..., [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - wat_a1_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- m
- long_name :
- aerosol water, interstitial, mode 01
- basename :
- wat_a1
array([[[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [3.573209e-10, 1.707779e-10, 3.096979e-10], [3.213501e-10, 1.530345e-10, 2.958907e-10]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [3.372876e-10, 1.803741e-10, 3.181657e-10], [3.049131e-10, 1.608551e-10, 3.030204e-10]], ..., [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [8.801577e-10, 9.321247e-10, 6.873683e-10], [7.739578e-10, 7.882232e-10, 5.932971e-10]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [8.511157e-10, 1.047754e-09, 7.255622e-10], [7.469533e-10, 8.792517e-10, 6.119186e-10]]], dtype=float32) - wat_a2_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- m
- long_name :
- aerosol water, interstitial, mode 02
- basename :
- wat_a2
array([[[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [4.221467e-12, 3.782512e-12, 7.559454e-12], [3.820588e-12, 3.399827e-12, 7.282526e-12]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [4.020182e-12, 4.074566e-12, 7.936143e-12], [3.655107e-12, 3.646458e-12, 7.638128e-12]], ..., [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [1.121921e-11, 1.397741e-11, 1.207264e-11], [9.977724e-12, 1.197653e-11, 1.056670e-11]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [1.101524e-11, 1.575757e-11, 1.188766e-11], [9.778303e-12, 1.341209e-11, 1.022033e-11]]], dtype=float32) - wat_a3_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- m
- long_name :
- aerosol water, interstitial, mode 03
- basename :
- wat_a3
array([[[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [2.897666e-09, 2.671686e-09, 4.835499e-09], [2.605710e-09, 2.395199e-09, 4.625470e-09]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [2.506672e-09, 2.872422e-09, 5.000213e-09], [2.265705e-09, 2.563129e-09, 4.763931e-09]], ..., [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [1.100520e-08, 3.887721e-09, 4.342406e-09], [9.662621e-09, 3.270303e-09, 3.727701e-09]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [1.102052e-08, 4.386234e-09, 4.263581e-09], [9.657203e-09, 3.661844e-09, 3.576182e-09]]], dtype=float32) - wat_a4_155w_to_158w_70n_to_73n(time, lev, ncol_155w_to_158w_70n_to_73n)float32...
- mdims :
- 9
- units :
- m
- long_name :
- aerosol water, interstitial, mode 04
- basename :
- wat_a4
array([[[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [1.918577e-17, 1.082542e-17, 1.359587e-17], [1.733146e-17, 9.714643e-18, 1.297123e-17]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [1.857251e-17, 1.250154e-17, 1.547616e-17], [1.685358e-17, 1.116824e-17, 1.471787e-17]], ..., [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [6.243886e-17, 5.984895e-17, 5.522160e-17], [5.526004e-17, 5.130619e-17, 4.807795e-17]], [[0.000000e+00, 0.000000e+00, 0.000000e+00], [0.000000e+00, 0.000000e+00, 0.000000e+00], ..., [7.555429e-17, 6.301745e-17, 6.484410e-17], [6.673959e-17, 5.367670e-17, 5.525207e-17]]], dtype=float32)
- Conventions :
- CF-1.0
- source :
- CAM
- case :
- EAMv1_ARM_Freerun_1
- title :
- UNSET
- logname :
- yshi
- host :
- cori07
- Version :
- $Name$
- revision_Id :
- $Id$
- initial_file :
- /global/cfs/cdirs/e3sm/inputdata/atm/cam/inic/homme/cami_mam3_Linoz_ne30np4_L72_c160214.nc
- topography_file :
- /global/cfs/cdirs/e3sm/inputdata/atm/cam/topo/USGS-gtopo30_ne30np4_16xdel2-PFC-consistentSGH.nc
- time_period_freq :
- hour_1
Instead of explicitly using the xarray package to load the data, EMC² can automatically load the data file when generating an emc2.core.model.E3SM object. By using EMC² to load a large-scale model’s output, we are able to handle 2 potential issues exemplified in the examined E3SM output file:
Because E3SM operates a cube-sphere grid, data is not provided in a strict lat-lon grid (two spatial dimensions) but rather on a column basis (one spatial dimension). We need to inform EMC² about that by setting
all_appended_in_lat=True.The regional output file has appended strings at the end of every field name, the result of post-processing machinery. By setting
appended_str=True, EMC² can remove these strings by using one of EMC²’s internal methods invoked during initialization.
Let’s generate an emc2.core.model.E3SM object and examine how the loaded data file is shown after initialization as an xr.Dataset object.
# Set model class object to use and load output file.
my_e3sm = emc2.core.model.E3SM(model_path, all_appended_in_lat=True, appended_str=True)
my_e3sm.ds # shows the loaded dataset
./data/EMC2_demo_EAMv1_Freerun.2012-10-31.nc is a regional output dataset; Stacking the time, lat, and lon dims for processing with EMC^2.
<xarray.Dataset>
Dimensions: (time_lat_lon: 72, lev: 72, cosp_prs: 7, nbnd: 2,
cosp_tau: 7, cosp_scol: 10, cosp_ht: 40, cosp_sr: 15,
cosp_sza: 5, cosp_htmisr: 16, cosp_tau_modis: 6,
ilev: 73, ncol_tmp: 3, time_tmp: 24)
Coordinates: (12/13)
* time_lat_lon (time_lat_lon) object (0, Timestamp('2012-10-31 01:0...
* lev (lev) float64 998.5 993.8 986.2 ... 0.1828 0.1238
* cosp_prs (cosp_prs) float64 900.0 740.0 620.0 ... 245.0 90.0
* cosp_tau (cosp_tau) float64 0.15 0.8 2.45 6.5 16.2 41.5 219.5
* cosp_scol (cosp_scol) int32 1 2 3 4 5 6 7 8 9 10
* cosp_ht (cosp_ht) float64 240.0 720.0 ... 1.848e+04 1.896e+04
... ...
* cosp_sza (cosp_sza) float64 0.0 15.0 30.0 45.0 60.0
* cosp_htmisr (cosp_htmisr) float64 -99.0 0.25 0.75 ... 16.0 58.0
* cosp_tau_modis (cosp_tau_modis) float64 0.8 2.45 ... 41.5 5.003e+04
* ilev (ilev) float64 0.1 0.1477 0.218 ... 990.5 997.0 1e+03
* ncol_tmp (ncol_tmp) int64 0 1 2
* time_tmp (time_tmp) datetime64[ns] 2012-10-31T01:00:00 ... 20...
Dimensions without coordinates: nbnd
Data variables: (12/209)
lat (time_lat_lon) float64 dask.array<chunksize=(72,), meta=np.ndarray>
lon (time_lat_lon) float64 dask.array<chunksize=(72,), meta=np.ndarray>
cosp_prs_bnds (cosp_prs, nbnd) float64 dask.array<chunksize=(7, 2), meta=np.ndarray>
cosp_tau_bnds (nbnd, cosp_tau) float64 dask.array<chunksize=(2, 7), meta=np.ndarray>
cosp_ht_bnds (nbnd, cosp_ht) float64 dask.array<chunksize=(2, 40), meta=np.ndarray>
cosp_sr_bnds (nbnd, cosp_sr) float64 dask.array<chunksize=(2, 15), meta=np.ndarray>
... ...
wat_a2 (time_lat_lon, lev) float32 3.821e-12 4.221e-12 ... 0.0
wat_a3 (time_lat_lon, lev) float32 2.606e-09 2.898e-09 ... 0.0
wat_a4 (time_lat_lon, lev) float32 1.733e-17 1.919e-17 ... 0.0
p_3d (time_lat_lon, lev) float64 dask.array<chunksize=(72, 72), meta=np.ndarray>
zeros_cf (time_lat_lon, lev) float64 0.0 0.0 0.0 ... 0.0 0.0 0.0
rho_a (time_lat_lon, lev) float64 dask.array<chunksize=(72, 72), meta=np.ndarray>
Attributes: (12/15)
Conventions: CF-1.0
source: CAM
case: EAMv1_ARM_Freerun_1
title: UNSET
logname: yshi
host: cori07
... ...
topography_file: /global/cfs/cdirs/e3sm/inputdata/atm/cam/topo/USGS-...
time_period_freq: hour_1
_file_dates: ['20121031']
_file_times: ['010000']
_datastream: act_datastream
_arm_standards_flag: 0- time_lat_lon: 72
- lev: 72
- cosp_prs: 7
- nbnd: 2
- cosp_tau: 7
- cosp_scol: 10
- cosp_ht: 40
- cosp_sr: 15
- cosp_sza: 5
- cosp_htmisr: 16
- cosp_tau_modis: 6
- ilev: 73
- ncol_tmp: 3
- time_tmp: 24
- time_lat_lon(time_lat_lon)object(0, Timestamp('2012-10-31 01:00:...
array([(0, Timestamp('2012-10-31 01:00:00')), (0, Timestamp('2012-10-31 02:00:00')), (0, Timestamp('2012-10-31 03:00:00')), (0, Timestamp('2012-10-31 04:00:00')), (0, Timestamp('2012-10-31 05:00:00')), (0, Timestamp('2012-10-31 06:00:00')), (0, Timestamp('2012-10-31 07:00:00')), (0, Timestamp('2012-10-31 08:00:00')), (0, Timestamp('2012-10-31 09:00:00')), (0, Timestamp('2012-10-31 10:00:00')), (0, Timestamp('2012-10-31 11:00:00')), (0, Timestamp('2012-10-31 12:00:00')), (0, Timestamp('2012-10-31 13:00:00')), (0, Timestamp('2012-10-31 14:00:00')), (0, Timestamp('2012-10-31 15:00:00')), (0, Timestamp('2012-10-31 16:00:00')), (0, Timestamp('2012-10-31 17:00:00')), (0, Timestamp('2012-10-31 18:00:00')), (0, Timestamp('2012-10-31 19:00:00')), (0, Timestamp('2012-10-31 20:00:00')), (0, Timestamp('2012-10-31 21:00:00')), (0, Timestamp('2012-10-31 22:00:00')), (0, Timestamp('2012-10-31 23:00:00')), (0, Timestamp('2012-11-01 00:00:00')), (1, Timestamp('2012-10-31 01:00:00')), (1, Timestamp('2012-10-31 02:00:00')), (1, Timestamp('2012-10-31 03:00:00')), (1, Timestamp('2012-10-31 04:00:00')), (1, Timestamp('2012-10-31 05:00:00')), (1, Timestamp('2012-10-31 06:00:00')), (1, Timestamp('2012-10-31 07:00:00')), (1, Timestamp('2012-10-31 08:00:00')), (1, Timestamp('2012-10-31 09:00:00')), (1, Timestamp('2012-10-31 10:00:00')), (1, Timestamp('2012-10-31 11:00:00')), (1, Timestamp('2012-10-31 12:00:00')), (1, Timestamp('2012-10-31 13:00:00')), (1, Timestamp('2012-10-31 14:00:00')), (1, Timestamp('2012-10-31 15:00:00')), (1, Timestamp('2012-10-31 16:00:00')), (1, Timestamp('2012-10-31 17:00:00')), (1, Timestamp('2012-10-31 18:00:00')), (1, Timestamp('2012-10-31 19:00:00')), (1, Timestamp('2012-10-31 20:00:00')), (1, Timestamp('2012-10-31 21:00:00')), (1, Timestamp('2012-10-31 22:00:00')), (1, Timestamp('2012-10-31 23:00:00')), (1, Timestamp('2012-11-01 00:00:00')), (2, Timestamp('2012-10-31 01:00:00')), (2, Timestamp('2012-10-31 02:00:00')), (2, Timestamp('2012-10-31 03:00:00')), (2, Timestamp('2012-10-31 04:00:00')), (2, Timestamp('2012-10-31 05:00:00')), (2, Timestamp('2012-10-31 06:00:00')), (2, Timestamp('2012-10-31 07:00:00')), (2, Timestamp('2012-10-31 08:00:00')), (2, Timestamp('2012-10-31 09:00:00')), (2, Timestamp('2012-10-31 10:00:00')), (2, Timestamp('2012-10-31 11:00:00')), (2, Timestamp('2012-10-31 12:00:00')), (2, Timestamp('2012-10-31 13:00:00')), (2, Timestamp('2012-10-31 14:00:00')), (2, Timestamp('2012-10-31 15:00:00')), (2, Timestamp('2012-10-31 16:00:00')), (2, Timestamp('2012-10-31 17:00:00')), (2, Timestamp('2012-10-31 18:00:00')), (2, Timestamp('2012-10-31 19:00:00')), (2, Timestamp('2012-10-31 20:00:00')), (2, Timestamp('2012-10-31 21:00:00')), (2, Timestamp('2012-10-31 22:00:00')), (2, Timestamp('2012-10-31 23:00:00')), (2, Timestamp('2012-11-01 00:00:00'))], dtype=object) - lev(lev)float64998.5 993.8 986.2 ... 0.1828 0.1238
array([9.984964e+02, 9.937570e+02, 9.862053e+02, 9.773141e+02, 9.679111e+02, 9.580128e+02, 9.476362e+02, 9.367990e+02, 9.255197e+02, 9.138175e+02, 9.017118e+02, 8.892227e+02, 8.763706e+02, 8.631764e+02, 8.496612e+02, 8.350952e+02, 8.178688e+02, 7.969527e+02, 7.723628e+02, 7.444748e+02, 7.137046e+02, 6.804980e+02, 6.453200e+02, 6.086438e+02, 5.706249e+02, 5.316395e+02, 4.924686e+02, 4.543958e+02, 4.188035e+02, 3.859990e+02, 3.557641e+02, 3.278975e+02, 3.022136e+02, 2.785416e+02, 2.567237e+02, 2.366148e+02, 2.180810e+02, 2.009989e+02, 1.852549e+02, 1.707441e+02, 1.573699e+02, 1.450433e+02, 1.336822e+02, 1.232110e+02, 1.135600e+02, 1.046650e+02, 9.646667e+01, 8.891054e+01, 8.194628e+01, 7.535534e+01, 6.847639e+01, 6.101518e+01, 5.330956e+01, 4.567120e+01, 3.836628e+01, 3.159325e+01, 2.544470e+01, 1.999634e+01, 1.533394e+01, 1.147379e+01, 8.377404e+00, 5.968449e+00, 4.129833e+00, 2.797027e+00, 1.894352e+00, 1.282995e+00, 8.689386e-01, 5.885091e-01, 3.985817e-01, 2.699489e-01, 1.828292e-01, 1.238254e-01]) - cosp_prs(cosp_prs)float64900.0 740.0 620.0 ... 245.0 90.0
- long_name :
- COSP Mean ISCCP pressure
- units :
- hPa
- bounds :
- cosp_prs_bnds
array([900., 740., 620., 500., 375., 245., 90.])
- cosp_tau(cosp_tau)float640.15 0.8 2.45 6.5 16.2 41.5 219.5
- long_name :
- COSP Mean ISCCP optical depth
- units :
- 1
- bounds :
- cosp_tau_bnds
array([1.500e-01, 8.000e-01, 2.450e+00, 6.500e+00, 1.620e+01, 4.150e+01, 2.195e+02]) - cosp_scol(cosp_scol)int321 2 3 4 5 6 7 8 9 10
- long_name :
- COSP subcolumn
array([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10], dtype=int32)
- cosp_ht(cosp_ht)float64240.0 720.0 ... 1.848e+04 1.896e+04
- long_name :
- COSP Mean Height for lidar and radar simulator outputs
- units :
- m
- bounds :
- cosp_ht_bnds
array([ 240., 720., 1200., 1680., 2160., 2640., 3120., 3600., 4080., 4560., 5040., 5520., 6000., 6480., 6960., 7440., 7920., 8400., 8880., 9360., 9840., 10320., 10800., 11280., 11760., 12240., 12720., 13200., 13680., 14160., 14640., 15120., 15600., 16080., 16560., 17040., 17520., 18000., 18480., 18960.]) - cosp_sr(cosp_sr)float640.605 2.1 4.0 ... 539.5 1.004e+03
- long_name :
- COSP Mean Scattering Ratio for lidar simulator CFAD output
- units :
- 1
- bounds :
- cosp_sr_bnds
array([6.050e-01, 2.100e+00, 4.000e+00, 6.000e+00, 8.500e+00, 1.250e+01, 1.750e+01, 2.250e+01, 2.750e+01, 3.500e+01, 4.500e+01, 5.500e+01, 7.000e+01, 5.395e+02, 1.004e+03]) - cosp_sza(cosp_sza)float640.0 15.0 30.0 45.0 60.0
- long_name :
- COSP Parasol SZA
- units :
- degrees
array([ 0., 15., 30., 45., 60.])
- cosp_htmisr(cosp_htmisr)float64-99.0 0.25 0.75 ... 14.0 16.0 58.0
- long_name :
- COSP MISR height
- units :
- km
- bounds :
- cosp_htmisr_bnds
array([-99. , 0.25, 0.75, 1.25, 1.75, 2.25, 2.75, 3.5 , 4.5 , 6. , 8. , 10. , 12. , 14. , 16. , 58. ]) - cosp_tau_modis(cosp_tau_modis)float640.8 2.45 6.5 16.2 41.5 5.003e+04
- long_name :
- COSP Mean MODIS optical depth
- units :
- 1
- bounds :
- cosp_tau_modis_bnds
array([8.000e-01, 2.450e+00, 6.500e+00, 1.620e+01, 4.150e+01, 5.003e+04])
- ilev(ilev)float640.1 0.1477 0.218 ... 997.0 1e+03
- long_name :
- hybrid level at interfaces (1000*(A+B))
- units :
- hPa
- positive :
- down
- standard_name :
- atmosphere_hybrid_sigma_pressure_coordinate
- formula_terms :
- a: hyai b: hybi p0: P0 ps: PS
array([1.000000e-01, 1.476508e-01, 2.180076e-01, 3.218901e-01, 4.752733e-01, 7.017450e-01, 1.036132e+00, 1.529858e+00, 2.258847e+00, 3.335207e+00, 4.924460e+00, 7.012439e+00, 9.742370e+00, 1.320520e+01, 1.746267e+01, 2.253000e+01, 2.835939e+01, 3.482711e+01, 4.190545e+01, 4.943694e+01, 5.718218e+01, 6.484818e+01, 7.210460e+01, 7.860608e+01, 8.528648e+01, 9.253461e+01, 1.003987e+02, 1.089312e+02, 1.181888e+02, 1.282332e+02, 1.391312e+02, 1.509554e+02, 1.637844e+02, 1.777038e+02, 1.928061e+02, 2.091918e+02, 2.269702e+02, 2.462594e+02, 2.671880e+02, 2.898952e+02, 3.145321e+02, 3.412629e+02, 3.702654e+02, 4.017327e+02, 4.358743e+02, 4.729174e+02, 5.120198e+02, 5.512593e+02, 5.899905e+02, 6.272970e+02, 6.633429e+02, 6.976532e+02, 7.297561e+02, 7.591936e+02, 7.855321e+02, 8.083734e+02, 8.273643e+02, 8.428261e+02, 8.564964e+02, 8.698564e+02, 8.828849e+02, 8.955606e+02, 9.078631e+02, 9.197720e+02, 9.312675e+02, 9.423305e+02, 9.529419e+02, 9.630837e+02, 9.727385e+02, 9.818896e+02, 9.905210e+02, 9.969929e+02, 1.000000e+03]) - ncol_tmp(ncol_tmp)int640 1 2
array([0, 1, 2])
- time_tmp(time_tmp)datetime64[ns]2012-10-31T01:00:00 ... 2012-11-01
array(['2012-10-31T01:00:00.000000000', '2012-10-31T02:00:00.000000000', '2012-10-31T03:00:00.000000000', '2012-10-31T04:00:00.000000000', '2012-10-31T05:00:00.000000000', '2012-10-31T06:00:00.000000000', '2012-10-31T07:00:00.000000000', '2012-10-31T08:00:00.000000000', '2012-10-31T09:00:00.000000000', '2012-10-31T10:00:00.000000000', '2012-10-31T11:00:00.000000000', '2012-10-31T12:00:00.000000000', '2012-10-31T13:00:00.000000000', '2012-10-31T14:00:00.000000000', '2012-10-31T15:00:00.000000000', '2012-10-31T16:00:00.000000000', '2012-10-31T17:00:00.000000000', '2012-10-31T18:00:00.000000000', '2012-10-31T19:00:00.000000000', '2012-10-31T20:00:00.000000000', '2012-10-31T21:00:00.000000000', '2012-10-31T22:00:00.000000000', '2012-10-31T23:00:00.000000000', '2012-11-01T00:00:00.000000000'], dtype='datetime64[ns]')
- lat(time_lat_lon)float64dask.array<chunksize=(72,), meta=np.ndarray>
- long_name :
- latitude
- units :
- degrees_north
Array Chunk Bytes 576 B 576 B Shape (72,) (72,) Count 5 Tasks 1 Chunks Type float64 numpy.ndarray - lon(time_lat_lon)float64dask.array<chunksize=(72,), meta=np.ndarray>
- long_name :
- longitude
- units :
- degrees_east
Array Chunk Bytes 576 B 576 B Shape (72,) (72,) Count 5 Tasks 1 Chunks Type float64 numpy.ndarray - cosp_prs_bnds(cosp_prs, nbnd)float64dask.array<chunksize=(7, 2), meta=np.ndarray>
Array Chunk Bytes 112 B 112 B Shape (7, 2) (7, 2) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - cosp_tau_bnds(nbnd, cosp_tau)float64dask.array<chunksize=(2, 7), meta=np.ndarray>
Array Chunk Bytes 112 B 112 B Shape (2, 7) (2, 7) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - cosp_ht_bnds(nbnd, cosp_ht)float64dask.array<chunksize=(2, 40), meta=np.ndarray>
Array Chunk Bytes 640 B 640 B Shape (2, 40) (2, 40) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - cosp_sr_bnds(nbnd, cosp_sr)float64dask.array<chunksize=(2, 15), meta=np.ndarray>
Array Chunk Bytes 240 B 240 B Shape (2, 15) (2, 15) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - cosp_htmisr_bnds(nbnd, cosp_htmisr)float64dask.array<chunksize=(2, 16), meta=np.ndarray>
Array Chunk Bytes 256 B 256 B Shape (2, 16) (2, 16) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - cosp_tau_modis_bnds(nbnd, cosp_tau_modis)float64dask.array<chunksize=(2, 6), meta=np.ndarray>
Array Chunk Bytes 96 B 96 B Shape (2, 6) (2, 6) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - hyam(lev)float640.0 0.0 ... 0.0001828 0.0001238
- long_name :
- hybrid A coefficient at layer midpoints
array([0.00000000e+00, 0.00000000e+00, 3.64808452e-04, 1.76556810e-03, 3.89449073e-03, 6.13557040e-03, 8.48494102e-03, 1.09385772e-02, 1.34923049e-02, 1.61418126e-02, 1.88826608e-02, 2.17103133e-02, 2.46201493e-02, 2.76074433e-02, 3.06674030e-02, 3.39653047e-02, 3.78655213e-02, 4.26011286e-02, 4.81685284e-02, 5.44826500e-02, 6.14493199e-02, 6.89676092e-02, 7.69322627e-02, 8.52361112e-02, 9.38439551e-02, 1.02670630e-01, 1.11539324e-01, 1.20159364e-01, 1.28217812e-01, 1.35645058e-01, 1.42490535e-01, 1.48799804e-01, 1.54614873e-01, 1.59974463e-01, 1.64914246e-01, 1.69467100e-01, 1.73663333e-01, 1.77530875e-01, 1.78544788e-01, 1.70744080e-01, 1.57369878e-01, 1.45043266e-01, 1.33682180e-01, 1.23210991e-01, 1.13560001e-01, 1.04664967e-01, 9.64666734e-02, 8.89105431e-02, 8.19462751e-02, 7.53553359e-02, 6.84763902e-02, 6.10151817e-02, 5.33095614e-02, 4.56711979e-02, 3.83662831e-02, 3.15932513e-02, 2.54446965e-02, 1.99963380e-02, 1.53339382e-02, 1.14737872e-02, 8.37740438e-03, 5.96844937e-03, 4.12983316e-03, 2.79702694e-03, 1.89435248e-03, 1.28299491e-03, 8.68938570e-04, 5.88509147e-04, 3.98581704e-04, 2.69948862e-04, 1.82829236e-04, 1.23825413e-04]) - hybm(lev)float640.9985 0.9938 0.9858 ... 0.0 0.0
- long_name :
- hybrid B coefficient at layer midpoints
array([0.99849644, 0.99375696, 0.98584052, 0.9755485 , 0.96401659, 0.9518772 , 0.93915122, 0.92586042, 0.91202744, 0.89767569, 0.88282917, 0.86751242, 0.8517505 , 0.83556898, 0.81899385, 0.80112987, 0.7800033 , 0.75435162, 0.72419431, 0.68999218, 0.65225532, 0.61153044, 0.56838771, 0.52340766, 0.47678095, 0.4289689 , 0.38092925, 0.33423645, 0.29058565, 0.25035397, 0.21327359, 0.1790977 , 0.14759877, 0.1185671 , 0.09180944, 0.06714769, 0.04441766, 0.02346807, 0.00671011, 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ]) - P0()float64...
- long_name :
- reference pressure
- units :
- Pa
array(100000.)
- hyai(ilev)float64dask.array<chunksize=(73,), meta=np.ndarray>
- long_name :
- hybrid A coefficient at layer interfaces
Array Chunk Bytes 584 B 584 B Shape (73,) (73,) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - hybi(ilev)float64dask.array<chunksize=(73,), meta=np.ndarray>
- long_name :
- hybrid B coefficient at layer interfaces
Array Chunk Bytes 584 B 584 B Shape (73,) (73,) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - date(time_lat_lon)int32dask.array<chunksize=(72,), meta=np.ndarray>
- long_name :
- current date (YYYYMMDD)
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 6 Tasks 1 Chunks Type int32 numpy.ndarray - datesec(time_lat_lon)int32dask.array<chunksize=(72,), meta=np.ndarray>
- long_name :
- current seconds of current date
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 6 Tasks 1 Chunks Type int32 numpy.ndarray - time_bnds(time_lat_lon, nbnd)objectdask.array<chunksize=(72, 2), meta=np.ndarray>
- long_name :
- time interval endpoints
Array Chunk Bytes 1.12 kiB 1.12 kiB Shape (72, 2) (72, 2) Count 7 Tasks 1 Chunks Type object numpy.ndarray - date_written(time_lat_lon)|S8dask.array<chunksize=(72,), meta=np.ndarray>
Array Chunk Bytes 576 B 576 B Shape (72,) (72,) Count 6 Tasks 1 Chunks Type |S8 numpy.ndarray - time_written(time_lat_lon)|S8dask.array<chunksize=(72,), meta=np.ndarray>
Array Chunk Bytes 576 B 576 B Shape (72,) (72,) Count 6 Tasks 1 Chunks Type |S8 numpy.ndarray - ndbase()int32...
- long_name :
- base day
array(0, dtype=int32)
- nsbase()int32...
- long_name :
- seconds of base day
array(0, dtype=int32)
- nbdate()int32...
- long_name :
- base date (YYYYMMDD)
array(20110101, dtype=int32)
- nbsec()int32...
- long_name :
- seconds of base date
array(0, dtype=int32)
- mdt()int32...
- long_name :
- timestep
- units :
- s
array(1800, dtype=int32)
- ndcur(time_lat_lon)int32dask.array<chunksize=(72,), meta=np.ndarray>
- long_name :
- current day (from base day)
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 6 Tasks 1 Chunks Type int32 numpy.ndarray - nscur(time_lat_lon)int32dask.array<chunksize=(72,), meta=np.ndarray>
- long_name :
- current seconds of current day
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 6 Tasks 1 Chunks Type int32 numpy.ndarray - co2vmr(time_lat_lon)float64dask.array<chunksize=(72,), meta=np.ndarray>
- long_name :
- co2 volume mixing ratio
Array Chunk Bytes 576 B 576 B Shape (72,) (72,) Count 6 Tasks 1 Chunks Type float64 numpy.ndarray - ch4vmr(time_lat_lon)float64dask.array<chunksize=(72,), meta=np.ndarray>
- long_name :
- ch4 volume mixing ratio
Array Chunk Bytes 576 B 576 B Shape (72,) (72,) Count 6 Tasks 1 Chunks Type float64 numpy.ndarray - n2ovmr(time_lat_lon)float64dask.array<chunksize=(72,), meta=np.ndarray>
- long_name :
- n2o volume mixing ratio
Array Chunk Bytes 576 B 576 B Shape (72,) (72,) Count 6 Tasks 1 Chunks Type float64 numpy.ndarray - f11vmr(time_lat_lon)float64dask.array<chunksize=(72,), meta=np.ndarray>
- long_name :
- f11 volume mixing ratio
Array Chunk Bytes 576 B 576 B Shape (72,) (72,) Count 6 Tasks 1 Chunks Type float64 numpy.ndarray - f12vmr(time_lat_lon)float64dask.array<chunksize=(72,), meta=np.ndarray>
- long_name :
- f12 volume mixing ratio
Array Chunk Bytes 576 B 576 B Shape (72,) (72,) Count 6 Tasks 1 Chunks Type float64 numpy.ndarray - sol_tsi(time_lat_lon)float64dask.array<chunksize=(72,), meta=np.ndarray>
- long_name :
- total solar irradiance
- units :
- W/m2
Array Chunk Bytes 576 B 576 B Shape (72,) (72,) Count 6 Tasks 1 Chunks Type float64 numpy.ndarray - nsteph(time_lat_lon)int32dask.array<chunksize=(72,), meta=np.ndarray>
- long_name :
- current timestep
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 6 Tasks 1 Chunks Type int32 numpy.ndarray - ACTNI(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- units :
- Micron
- long_name :
- Average Cloud Top ice number
- basename :
- ACTNI
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - ACTNL(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- units :
- Micron
- long_name :
- Average Cloud Top droplet number
- basename :
- ACTNL
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - ACTREI(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- units :
- Micron
- long_name :
- Average Cloud Top ice effective radius
- basename :
- ACTREI
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - ACTREL(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- units :
- Micron
- long_name :
- Average Cloud Top droplet effective radius
- basename :
- ACTREL
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - ADRAIN(time_lat_lon, lev)float3255.72 42.64 29.77 ... nan nan nan
- mdims :
- 9
- units :
- Micron
- long_name :
- Average rain effective Diameter
- basename :
- ADRAIN
array([[55.72431 , 42.64484 , 29.769634, ..., nan, nan, nan], [54.873734, 41.86107 , 29.126762, ..., nan, nan, nan], [45.75345 , 43.76058 , 33.783154, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [56.918602, 48.124565, 37.298603, ..., nan, nan, nan]], dtype=float32) - ADSNOW(time_lat_lon, lev)float32nan nan nan nan ... nan nan nan nan
- mdims :
- 9
- units :
- Micron
- long_name :
- Average snow effective Diameter
- basename :
- ADSNOW
array([[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [1316.5657 , 1107.3442 , 854.9008 , ..., nan, nan, nan], [1223.0029 , 1027.803 , 792.3841 , ..., nan, nan, nan], [ 437.46613, 403.1429 , 354.79236, ..., nan, nan, nan]], dtype=float32) - ANRAIN(time_lat_lon, lev)float320.7012 1.03 3.508 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- m-3
- long_name :
- Average rain number conc
- basename :
- ANRAIN
array([[0.7011616 , 1.0301039 , 3.5078678 , ..., 0. , 0. , 0. ], [0.5778354 , 0.8504793 , 2.9005358 , ..., 0. , 0. , 0. ], [0.15508625, 0.37922254, 1.125233 , ..., 0. , 0. , 0. ], ..., [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0.00674652, ..., 0. , 0. , 0. ], [0.08697851, 0.1384414 , 0.29919252, ..., 0. , 0. , 0. ]], dtype=float32) - ANSNOW(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- m-3
- long_name :
- Average snow number conc
- basename :
- ANSNOW
array([[0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], [0. , 0. , 0. , ..., 0. , 0. , 0. ], ..., [0.00078325, 0.00110998, 0.00200698, ..., 0. , 0. , 0. ], [0.00111829, 0.00156309, 0.00275171, ..., 0. , 0. , 0. ], [0.01632102, 0.01890629, 0.024763 , ..., 0. , 0. , 0. ]], dtype=float32) - AODVIS(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- long_name :
- Aerosol optical depth 550 nm
- basename :
- AODVIS
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - AQRAIN(time_lat_lon, lev)float321.48e-10 1.914e-10 ... 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Average rain mixing ratio
- basename :
- AQRAIN
array([[1.4796600e-10, 1.9144121e-10, 5.6471416e-10, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [1.1400540e-10, 1.4737818e-10, 4.3401210e-10, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [1.9047700e-10, 2.4701019e-10, 3.5778944e-10, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 1.3524976e-11, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [1.7780903e-10, 1.9897498e-10, 2.4917188e-10, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], dtype=float32) - AQSNOW(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Average snow mixing ratio
- basename :
- AQSNOW
array([[0.0000000e+00, 0.0000000e+00, 0.0000000e+00, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [4.5607993e-09, 4.5855728e-09, 4.6392388e-09, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [5.4650018e-09, 5.4773408e-09, 5.5014517e-09, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [5.7880962e-09, 5.7902074e-09, 5.7977103e-09, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], dtype=float32) - AREI(time_lat_lon, lev)float32nan nan nan nan ... nan nan nan nan
- mdims :
- 9
- units :
- Micron
- long_name :
- Average ice effective radius
- basename :
- AREI
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - AREL(time_lat_lon, lev)float32nan nan nan nan ... nan nan nan nan
- mdims :
- 9
- units :
- Micron
- long_name :
- Average droplet effective radius
- basename :
- AREL
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - AWNC(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- m-3
- long_name :
- Average cloud water number conc
- basename :
- AWNC
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - AWNI(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- m-3
- long_name :
- Average cloud ice number conc
- basename :
- AWNI
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - CCN3(time_lat_lon, lev)float3210.09 10.03 9.932 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- #/cm3
- long_name :
- CCN concentration at S=0.1%
- basename :
- CCN3
array([[10.087448, 10.027431, 9.931998, ..., 0. , 0. , 0. ], [10.045017, 9.983613, 9.885693, ..., 0. , 0. , 0. ], [10.231188, 10.16933 , 10.07049 , ..., 0. , 0. , 0. ], ..., [14.040397, 13.94825 , 13.780395, ..., 0. , 0. , 0. ], [13.983971, 13.857922, 13.595864, ..., 0. , 0. , 0. ], [13.140232, 12.998683, 12.686504, ..., 0. , 0. , 0. ]], dtype=float32) - CDNUMC(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- units :
- 1/m2
- long_name :
- Vertically-integrated droplet concentration
- basename :
- CDNUMC
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - CLDHGH(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- units :
- fraction
- long_name :
- Vertically-integrated high cloud
- basename :
- CLDHGH
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - CLDICE(time_lat_lon, lev)float324.923e-15 1.902e-13 ... 3.773e-24
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Grid box averaged cloud ice amount
- basename :
- CLDICE
array([[4.9226556e-15, 1.9024746e-13, 8.1440887e-12, ..., 4.2775994e-24, 3.8617147e-24, 3.6965450e-24], [2.9488481e-16, 1.5516187e-14, 9.2084413e-13, ..., 4.2435226e-24, 3.8657458e-24, 3.7516308e-24], [8.2111417e-13, 7.5937802e-13, 8.6103596e-13, ..., 4.2540870e-24, 3.8876458e-24, 3.8097623e-24], ..., [1.6518245e-16, 2.0013909e-16, 5.6420976e-16, ..., 4.2676480e-24, 3.9434167e-24, 3.6891459e-24], [2.6899510e-14, 2.4054153e-14, 9.1016053e-15, ..., 4.2977742e-24, 3.9374107e-24, 3.7131170e-24], [5.5393759e-14, 3.7547788e-14, 1.1006347e-14, ..., 4.3099759e-24, 3.8853735e-24, 3.7734672e-24]], dtype=float32) - CLDLIQ(time_lat_lon, lev)float324.305e-36 1.99e-35 ... 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Grid box averaged cloud liquid amount
- basename :
- CLDLIQ
array([[4.3050988e-36, 1.9900146e-35, 5.9071848e-22, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [8.2156167e-38, 8.8877467e-38, 6.4625698e-31, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 1.2640355e-34, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 2.3869881e-18, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], dtype=float32) - CLDLOW(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- units :
- fraction
- long_name :
- Vertically-integrated low cloud
- basename :
- CLDLOW
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - CLDMED(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- units :
- fraction
- long_name :
- Vertically-integrated mid-level cloud
- basename :
- CLDMED
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - CLDTOT(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- units :
- fraction
- long_name :
- Vertically-integrated total cloud
- basename :
- CLDTOT
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - CLOUD(time_lat_lon, lev)float322.374e-05 6.296e-05 ... 0.0 0.0
- mdims :
- 9
- units :
- fraction
- long_name :
- Cloud fraction
- basename :
- CLOUD
array([[2.3736337e-05, 6.2959516e-05, 1.4467775e-04, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [2.2758020e-05, 6.4255721e-05, 1.4255832e-04, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 1.6439892e-05, 4.7910871e-05, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], dtype=float32) - CMFDICE(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg/s
- long_name :
- Cloud ice tendency - shallow convection
- basename :
- CMFDICE
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - CMFDLIQ(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg/s
- long_name :
- Cloud liq tendency - shallow convection
- basename :
- CMFDLIQ
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - CMFDQ(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg/s
- long_name :
- QV tendency - shallow convection
- basename :
- CMFDQ
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - CMFDT(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- K/s
- long_name :
- T tendency - shallow convection
- basename :
- CMFDT
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - CONCLD(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- fraction
- long_name :
- Convective cloud cover
- basename :
- CONCLD
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - EMIS(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- 1
- long_name :
- Cloud longwave emissivity
- basename :
- EMIS
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - EVAPPREC(time_lat_lon, lev)float324.923e-09 6.371e-09 ... 0.0 0.0
- mdims :
- 9
- units :
- kg/kg/s
- long_name :
- Rate of evaporation of falling precip
- basename :
- EVAPPREC
array([[4.92341634e-09, 6.37081854e-09, 1.29959918e-08, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [3.79439591e-09, 4.90591656e-09, 1.01390505e-08, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [5.91090288e-09, 8.22080004e-09, 1.19149490e-08, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], ..., [1.70121556e-10, 1.51254814e-10, 1.01600450e-10, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [1.85027701e-10, 1.47808349e-10, 5.21626180e-11, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [7.99513122e-10, 5.81052872e-10, 1.80768220e-10, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], dtype=float32) - EVAPSNOW(time_lat_lon, lev)float324.803e-13 3.831e-13 ... 0.0 0.0
- mdims :
- 9
- units :
- kg/kg/s
- long_name :
- Rate of evaporation of falling snow
- basename :
- EVAPSNOW
array([[4.8033084e-13, 3.8311742e-13, 2.4037814e-13, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [3.9146580e-13, 3.1312838e-13, 1.9882952e-13, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [3.0213123e-13, 2.4635526e-13, 1.6122047e-13, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [1.6995949e-10, 1.5098180e-10, 1.0103474e-10, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [1.8313948e-10, 1.4540184e-10, 4.8753685e-11, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [7.6169632e-10, 5.2378046e-10, 7.0503305e-11, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], dtype=float32) - FCTI(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- units :
- fraction
- long_name :
- Fractional occurrence of cloud top ice
- basename :
- FCTI
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - FCTL(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- units :
- fraction
- long_name :
- Fractional occurrence of cloud top liquid
- basename :
- FCTL
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - FDL(time_lat_lon, ilev)float32dask.array<chunksize=(72, 73), meta=np.ndarray>
- mdims :
- 10
- units :
- W/m2
- long_name :
- Longwave downward flux
- basename :
- FDL
Array Chunk Bytes 20.53 kiB 20.53 kiB Shape (72, 73) (72, 73) Count 5 Tasks 1 Chunks Type float32 numpy.ndarray - FDLC(time_lat_lon, ilev)float32dask.array<chunksize=(72, 73), meta=np.ndarray>
- mdims :
- 10
- units :
- W/m2
- long_name :
- Longwave clear-sky downward flux
- basename :
- FDLC
Array Chunk Bytes 20.53 kiB 20.53 kiB Shape (72, 73) (72, 73) Count 5 Tasks 1 Chunks Type float32 numpy.ndarray - FDS(time_lat_lon, ilev)float32dask.array<chunksize=(72, 73), meta=np.ndarray>
- mdims :
- 10
- units :
- W/m2
- long_name :
- Shortwave downward flux
- basename :
- FDS
Array Chunk Bytes 20.53 kiB 20.53 kiB Shape (72, 73) (72, 73) Count 5 Tasks 1 Chunks Type float32 numpy.ndarray - FDSC(time_lat_lon, ilev)float32dask.array<chunksize=(72, 73), meta=np.ndarray>
- mdims :
- 10
- units :
- W/m2
- long_name :
- Shortwave clear-sky downward flux
- basename :
- FDSC
Array Chunk Bytes 20.53 kiB 20.53 kiB Shape (72, 73) (72, 73) Count 5 Tasks 1 Chunks Type float32 numpy.ndarray - FICE(time_lat_lon, lev)float320.001445 0.001034 ... 0.0 0.0
- mdims :
- 9
- units :
- fraction
- long_name :
- Fractional ice content within cloud
- basename :
- FICE
array([[1.4454841e-03, 1.0339771e-03, 4.5222286e-04, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [1.5939100e-03, 1.1151755e-03, 4.5741317e-04, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [7.2889542e-04, 5.0388201e-04, 2.9065495e-04, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [9.9997902e-01, 9.9997032e-01, 9.9994421e-01, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [9.9849594e-01, 9.9822980e-01, 9.9754757e-01, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [9.7019577e-01, 9.6677762e-01, 9.5879334e-01, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], dtype=float32) - FLDS(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Downwelling longwave flux at surface
- basename :
- FLDS
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - FLDSC(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Clearsky Downwelling longwave flux at surface
- basename :
- FLDSC
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - FLN200(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Net longwave flux at 200 mb
- basename :
- FLN200
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - FLNS(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Net longwave flux at surface
- basename :
- FLNS
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - FLNT(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Net longwave flux at top of model
- basename :
- FLNT
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - FLUT(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Upwelling longwave flux at top of model
- basename :
- FLUT
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - FLUTC(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Clearsky upwelling longwave flux at top of model
- basename :
- FLUTC
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - FREQI(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- fraction
- long_name :
- Fractional occurrence of ice
- basename :
- FREQI
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - FREQL(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- fraction
- long_name :
- Fractional occurrence of liquid
- basename :
- FREQL
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - FREQR(time_lat_lon, lev)float320.0001 0.0001 0.0001447 ... 0.0 0.0
- mdims :
- 9
- units :
- fraction
- long_name :
- Fractional occurrence of rain
- basename :
- FREQR
array([[9.9999997e-05, 9.9999997e-05, 1.4467775e-04, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [9.9999997e-05, 9.9999997e-05, 1.4255832e-04, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [9.9999997e-05, 9.9999997e-05, 9.9999997e-05, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 9.9999997e-05, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [9.9999997e-05, 9.9999997e-05, 9.9999997e-05, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], dtype=float32) - FREQS(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- fraction
- long_name :
- Fractional occurrence of snow
- basename :
- FREQS
array([[0.e+00, 0.e+00, 0.e+00, ..., 0.e+00, 0.e+00, 0.e+00], [0.e+00, 0.e+00, 0.e+00, ..., 0.e+00, 0.e+00, 0.e+00], [0.e+00, 0.e+00, 0.e+00, ..., 0.e+00, 0.e+00, 0.e+00], ..., [1.e-04, 1.e-04, 1.e-04, ..., 0.e+00, 0.e+00, 0.e+00], [1.e-04, 1.e-04, 1.e-04, ..., 0.e+00, 0.e+00, 0.e+00], [1.e-04, 1.e-04, 1.e-04, ..., 0.e+00, 0.e+00, 0.e+00]], dtype=float32) - FSDS(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Downwelling solar flux at surface
- basename :
- FSDS
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - FSDSC(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Clearsky downwelling solar flux at surface
- basename :
- FSDSC
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - FSNS(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Net solar flux at surface
- basename :
- FSNS
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - FSNT(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Net solar flux at top of model
- basename :
- FSNT
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - FSNTOAC(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Clearsky net solar flux at top of atmosphere
- basename :
- FSNTOAC
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - FSUTOA(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Upwelling solar flux at top of atmosphere
- basename :
- FSUTOA
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - GCLDLWP(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/m2
- long_name :
- Grid-box cloud water path
- basename :
- GCLDLWP
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - ICE_ICLD_VISTAU(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- Sampling_Sequence :
- rad_lwsw
- units :
- 1
- long_name :
- Ice in-cloud extinction visible sw optical depth
- basename :
- ICE_ICLD_VISTAU
array([[ 0., 0., 0., ..., 0., 0., 0.], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [ 0., 0., 0., ..., 0., 0., 0.], [ 0., 0., 0., ..., 0., 0., 0.], [ 0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - ICIMR(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Prognostic in-cloud ice mixing ratio
- basename :
- ICIMR
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - ICIMRST(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Prognostic in-stratus ice mixing ratio
- basename :
- ICIMRST
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - ICINC(time_lat_lon, lev)float321.442 5.448 7.747 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- m-3
- long_name :
- Prognostic in-cloud ice number conc
- basename :
- ICINC
array([[1.4422982e+00, 5.4483218e+00, 7.7469735e+00, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [1.0096334e+00, 4.0283918e+00, 5.0887146e+00, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [1.9913820e+00, 2.0800712e+00, 2.8134835e+00, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [9.4607944e-04, 1.3059836e-03, 4.9493276e-03, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [3.3250146e-02, 3.7625652e-02, 2.6070204e-02, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [9.6784895e-03, 1.7518008e-02, 4.3427676e-02, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], dtype=float32) - ICLDIWP(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/m2
- long_name :
- In-cloud ice water path
- basename :
- ICLDIWP
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - ICLDTWP(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/m2
- long_name :
- In-cloud cloud total water path (liquid and ice)
- basename :
- ICLDTWP
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - ICWMRST(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Prognostic in-stratus water mixing ratio
- basename :
- ICWMRST
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - ICWNC(time_lat_lon, lev)float321.347e-08 1.343e-08 ... 0.0 0.0
- mdims :
- 9
- units :
- m-3
- long_name :
- Prognostic in-cloud water number conc
- basename :
- ICWNC
array([[1.34728992e-08, 1.34277940e-08, 9.23126020e-09, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [1.34724161e-08, 1.34271447e-08, 9.36786382e-09, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [1.34719711e-08, 1.34265061e-08, 1.33537785e-08, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], ..., [1.33342573e-08, 1.32865043e-08, 1.32080382e-08, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [1.32836417e-08, 1.32369387e-08, 1.31608235e-08, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [1.32281697e-08, 1.31819435e-08, 2.29554325e-02, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], dtype=float32) - IWC(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/m3
- long_name :
- Grid box average ice water content
- basename :
- IWC
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - LIQ_ICLD_VISTAU(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- Sampling_Sequence :
- rad_lwsw
- units :
- 1
- long_name :
- Liquid in-cloud extinction visible sw optical depth
- basename :
- LIQ_ICLD_VISTAU
array([[ 0., 0., 0., ..., 0., 0., 0.], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [ 0., 0., 0., ..., 0., 0., 0.], [ 0., 0., 0., ..., 0., 0., 0.], [ 0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - LWC(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/m3
- long_name :
- Grid box average liquid water content
- basename :
- LWC
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - LWCF(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Longwave cloud forcing
- basename :
- LWCF
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - NSNOW(time_lat_lon, lev)float320.009382 0.009571 ... 0.0 0.0
- mdims :
- 9
- units :
- m-3
- long_name :
- Diagnostic grid-mean snow number conc
- basename :
- NSNOW
array([[9.38184839e-03, 9.57082864e-03, 1.00646671e-02, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [6.50361134e-03, 6.65427418e-03, 6.99624699e-03, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [4.33164136e-03, 4.50564316e-03, 4.73473966e-03, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], ..., [7.83252907e+00, 1.10998278e+01, 2.00697708e+01, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [1.11828718e+01, 1.56308565e+01, 2.75170879e+01, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [1.63210236e+02, 1.89062881e+02, 2.47630005e+02, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], dtype=float32) - NUMICE(time_lat_lon, lev)float321.443e-10 5.45e-10 ... 1.627e-22
- mdims :
- 9
- units :
- 1/kg
- long_name :
- Grid box averaged cloud ice number
- basename :
- NUMICE
array([[1.4427604e-10, 5.4500676e-10, 1.1211739e-09, ..., 3.6240230e-22, 2.3979228e-22, 1.6293956e-22], [1.0099570e-10, 4.0296827e-10, 7.2567113e-10, ..., 3.6164436e-22, 2.3812298e-22, 1.6258192e-22], [1.9920202e-10, 2.0807377e-10, 2.8143854e-10, ..., 3.6153394e-22, 2.3840929e-22, 1.6324809e-22], ..., [9.4638253e-14, 1.3064020e-13, 4.9509133e-13, ..., 3.6758040e-22, 2.4783059e-22, 1.6340139e-22], [3.3260801e-12, 3.7637710e-12, 2.6078556e-12, ..., 3.6765932e-22, 2.4619417e-22, 1.6339610e-22], [9.6815904e-13, 1.7523620e-12, 4.3441591e-12, ..., 3.6622339e-22, 2.4382497e-22, 1.6272291e-22]], dtype=float32) - NUMIMM10sBC(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- #/m3
- long_name :
- Ice Number Concentration due to imm freezing by bc in Mixed Clouds during 10-s period
- basename :
- NUMIMM10sBC
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - NUMIMM10sDST(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- #/m3
- long_name :
- Ice Number Concentration due to imm freezing by dst in Mixed Clouds during 10-s period
- basename :
- NUMIMM10sDST
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - NUMLIQ(time_lat_lon, lev)float321.348e-18 1.343e-18 ... 1.627e-22
- mdims :
- 9
- units :
- 1/kg
- long_name :
- Grid box averaged cloud liquid number
- basename :
- NUMLIQ
array([[1.3477215e-18, 1.3432097e-18, 1.3359860e-18, ..., 3.6240230e-22, 2.3979228e-22, 1.6293956e-22], [1.3476734e-18, 1.3431448e-18, 1.3358949e-18, ..., 3.6164436e-22, 2.3812298e-22, 1.6258192e-22], [1.3476288e-18, 1.3430808e-18, 1.3358058e-18, ..., 3.6153394e-22, 2.3840929e-22, 1.6324809e-22], ..., [1.3338529e-18, 1.3290761e-18, 1.3212270e-18, ..., 3.6758040e-22, 2.4783059e-22, 1.6340139e-22], [1.3287898e-18, 1.3241181e-18, 1.3165041e-18, ..., 3.6765932e-22, 2.4619417e-22, 1.6339610e-22], [1.3232408e-18, 1.3186167e-18, 2.2962786e-12, ..., 3.6622339e-22, 2.4382497e-22, 1.6272291e-22]], dtype=float32) - NUMRAI(time_lat_lon, lev)float320.001946 0.004853 ... 1.627e-22
- mdims :
- 9
- units :
- 1/kg
- long_name :
- Grid box averaged rain number
- basename :
- NUMRAI
array([[1.94640318e-03, 4.85288352e-03, 1.68888103e-02, ..., 3.62402296e-22, 2.39792283e-22, 1.62939561e-22], [1.59615313e-03, 4.03811364e-03, 1.43019333e-02, ..., 3.61644358e-22, 2.38122978e-22, 1.62581923e-22], [2.02719573e-04, 1.21054635e-03, 4.09822864e-03, ..., 3.61533943e-22, 2.38409290e-22, 1.63248087e-22], ..., [1.00208837e-07, 3.26158215e-07, 1.79881943e-06, ..., 3.67580404e-22, 2.47830587e-22, 1.63401391e-22], [4.49929712e-06, 1.00217485e-05, 3.60330196e-05, ..., 3.67659316e-22, 2.46194174e-22, 1.63396103e-22], [4.37628973e-04, 8.62879097e-04, 2.51152948e-03, ..., 3.66223387e-22, 2.43824966e-22, 1.62722908e-22]], dtype=float32) - NUMSNO(time_lat_lon, lev)float321.137e-08 1.146e-08 ... 1.627e-22
- mdims :
- 9
- units :
- 1/kg
- long_name :
- Grid box averaged snow number
- basename :
- NUMSNO
array([[1.1368726e-08, 1.1456228e-08, 1.1541645e-08, ..., 3.6240230e-22, 2.3979228e-22, 1.6293956e-22], [7.9196596e-09, 8.0496401e-09, 8.2013223e-09, ..., 3.6164436e-22, 2.3812298e-22, 1.6258192e-22], [4.7537139e-09, 5.1927502e-09, 5.4241114e-09, ..., 3.6153394e-22, 2.3840929e-22, 1.6324809e-22], ..., [4.3351010e-06, 7.2621933e-06, 1.5678894e-05, ..., 3.6758040e-22, 2.4783059e-22, 1.6340139e-22], [6.2836448e-06, 1.0544682e-05, 2.2833710e-05, ..., 3.6765932e-22, 2.4619417e-22, 1.6339610e-22], [1.5086352e-04, 1.9219190e-04, 2.8078773e-04, ..., 3.6622339e-22, 2.4382497e-22, 1.6272291e-22]], dtype=float32) - OMEGA(time_lat_lon, lev)float320.01072 0.01648 ... 5.859e-05
- mdims :
- 9
- units :
- Pa/s
- long_name :
- Vertical velocity (pressure)
- basename :
- OMEGA
array([[ 1.07247764e-02, 1.64787862e-02, 2.51672845e-02, ..., 1.54458408e-04, 2.00939015e-04, 9.20130624e-05], [ 6.95641851e-03, 1.31674763e-02, 2.25521848e-02, ..., 1.15136179e-04, 1.23753503e-04, 5.22241717e-05], [ 1.33657139e-02, 1.99950822e-02, 3.00504137e-02, ..., 2.98087652e-05, 4.08378291e-05, 3.73170660e-05], ..., [ 6.15722053e-02, 6.88638687e-02, 6.11765236e-02, ..., 1.50428139e-04, 4.49113759e-05, -8.57438772e-06], [ 6.21933192e-02, 6.34866282e-02, 5.00574708e-02, ..., 1.77030641e-04, 1.15848736e-04, 2.73728801e-05], [ 5.49512506e-02, 4.96051274e-02, 3.00648753e-02, ..., 1.45226717e-04, 1.52054767e-04, 5.85891103e-05]], dtype=float32) - PRAO(time_lat_lon, lev)float320.0 0.0 0.0 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg/s
- long_name :
- Accretion of cloud water by rain
- basename :
- PRAO
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - PRCO(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg/s
- long_name :
- Autoconversion of cloud water
- basename :
- PRCO
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - PRECC(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- units :
- m/s
- long_name :
- Convective precipitation rate (liq + ice)
- basename :
- PRECC
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - PRECL(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- units :
- m/s
- long_name :
- Large-scale (stable) precipitation rate (liq + ice)
- basename :
- PRECL
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - PRECT(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- units :
- m/s
- long_name :
- Total (convective and large-scale) precipitation rate (liq + ice)
- basename :
- PRECT
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - PS(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- units :
- Pa
- long_name :
- Surface pressure
- basename :
- PS
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - PSL(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- units :
- Pa
- long_name :
- Sea level pressure
- basename :
- PSL
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - Q(time_lat_lon, lev)float320.001912 0.001905 ... 2.172e-06
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Specific humidity
- basename :
- Q
array([[1.9123822e-03, 1.9053145e-03, 1.8975340e-03, ..., 2.1726269e-06, 2.1540654e-06, 2.1281078e-06], [1.9103336e-03, 1.9034640e-03, 1.8959978e-03, ..., 2.1690369e-06, 2.1528379e-06, 2.1399544e-06], [1.9023425e-03, 1.8965562e-03, 1.8902243e-03, ..., 2.1676826e-06, 2.1551232e-06, 2.1503724e-06], ..., [1.9106519e-03, 1.9055578e-03, 1.9003357e-03, ..., 2.1620565e-06, 2.1655483e-06, 2.1797139e-06], [2.0498680e-03, 2.0459704e-03, 2.0416782e-03, ..., 2.1608093e-06, 2.1642998e-06, 2.1773180e-06], [2.2063514e-03, 2.2037136e-03, 2.2012063e-03, ..., 2.1627166e-06, 2.1664714e-06, 2.1724954e-06]], dtype=float32) - QRAIN(time_lat_lon, lev)float321.48e-06 1.914e-06 ... 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Diagnostic grid-mean rain mixing ratio
- basename :
- QRAIN
array([[1.4796600e-06, 1.9144120e-06, 3.9032552e-06, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [1.1400541e-06, 1.4737818e-06, 3.0444530e-06, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [1.9047699e-06, 2.4701019e-06, 3.5778944e-06, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [9.5697317e-10, 1.3609546e-09, 2.5890403e-09, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [8.2322373e-08, 9.7132769e-08, 1.3524976e-07, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [1.7780903e-06, 1.9897498e-06, 2.4917188e-06, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], dtype=float32) - QSNOW(time_lat_lon, lev)float322.142e-09 1.981e-09 ... 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Diagnostic grid-mean snow mixing ratio
- basename :
- QSNOW
array([[2.1419162e-09, 1.9813169e-09, 1.7577956e-09, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [1.8200443e-09, 1.6453449e-09, 1.3922893e-09, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [1.3885696e-09, 1.2445081e-09, 1.0393739e-09, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [4.5607994e-05, 4.5855730e-05, 4.6392386e-05, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [5.4650016e-05, 5.4773409e-05, 5.5014516e-05, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [5.7880963e-05, 5.7902074e-05, 5.7977104e-05, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], dtype=float32) - QVRES(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg/s
- long_name :
- Rate of residual condensation term
- basename :
- QVRES
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - RAINQM(time_lat_lon, lev)float326.281e-06 7.042e-06 ... 1.202e-24
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Grid box averaged rain amount
- basename :
- RAINQM
array([[6.2806789e-06, 7.0419896e-06, 8.3821915e-06, ..., 1.1930321e-24, 1.2329296e-24, 1.2767012e-24], [4.9183936e-06, 5.5427658e-06, 6.6487332e-06, ..., 1.1880765e-24, 1.2509350e-24, 1.2633745e-24], [3.6210685e-07, 1.8983151e-06, 2.9729897e-06, ..., 1.1937434e-24, 1.2303874e-24, 1.2189339e-24], ..., [2.5292965e-10, 4.3386070e-10, 1.0624546e-09, ..., 1.1178710e-24, 1.1588297e-24, 1.2672579e-24], [3.6680504e-08, 4.7371099e-08, 7.4915931e-08, ..., 1.1145571e-24, 1.1495750e-24, 1.2418144e-24], [1.5327441e-06, 1.8330389e-06, 2.4981409e-06, ..., 1.1067339e-24, 1.1648636e-24, 1.2020652e-24]], dtype=float32) - RELHUM(time_lat_lon, lev)float3283.73 85.61 ... 0.0002166 0.0002172
- mdims :
- 9
- units :
- percent
- long_name :
- Relative humidity
- basename :
- RELHUM
array([[8.3730804e+01, 8.5614693e+01, 8.8982658e+01, ..., 2.1726268e-04, 2.1540654e-04, 2.1281077e-04], [8.3065056e+01, 8.4900948e+01, 8.8228500e+01, ..., 2.1690367e-04, 2.1528378e-04, 2.1399544e-04], [8.2156151e+01, 8.3978065e+01, 8.7271988e+01, ..., 2.1676825e-04, 2.1551231e-04, 2.1503725e-04], ..., [8.8453629e+01, 9.0072647e+01, 9.2647179e+01, ..., 2.1620565e-04, 2.1655482e-04, 2.1797139e-04], [8.9606087e+01, 9.1476593e+01, 9.4459862e+01, ..., 2.1608094e-04, 2.1642998e-04, 2.1773181e-04], [9.0189697e+01, 9.2181778e+01, 9.5297989e+01, ..., 2.1627167e-04, 2.1664714e-04, 2.1724954e-04]], dtype=float32) - RERCLD(time_lat_lon, lev)float325.911e-05 5.194e-05 ... 0.0 0.0
- mdims :
- 9
- units :
- m
- long_name :
- Diagnostic effective radius of Liquid Cloud and Rain
- basename :
- RERCLD
array([[5.91073149e-05, 5.19358910e-05, 4.40789590e-05, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [6.30436261e-05, 5.53605314e-05, 4.65019839e-05, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [9.77340314e-05, 7.24628189e-05, 5.03359188e-05, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], ..., [1.21731464e-04, 1.05471401e-04, 8.77150815e-05, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [1.97550704e-04, 1.72877451e-04, 1.41538781e-04, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [1.17794450e-04, 1.00770529e-04, 7.77939204e-05, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], dtype=float32) - RHCFMIP(time_lat_lon, lev)float3287.35 89.46 ... 1.815e-05 8.739e-06
- mdims :
- 9
- units :
- percent
- long_name :
- Relative humidity with respect to water above 273 K, ice below 273 K
- basename :
- RHCFMIP
array([[8.73468475e+01, 8.94625854e+01, 9.31331100e+01, ..., 3.14275967e-05, 1.22271085e-05, 8.82900440e-06], [8.66067123e+01, 8.86868210e+01, 9.22966232e+01, ..., 2.98583691e-05, 1.04015999e-05, 8.43694488e-06], [8.56140823e+01, 8.76887665e+01, 9.12698364e+01, ..., 2.96251437e-05, 1.07044179e-05, 9.32243165e-06], ..., [9.25081711e+01, 9.43432846e+01, 9.71473083e+01, ..., 4.38797069e-05, 2.67102241e-05, 9.65839445e-06], [9.35536499e+01, 9.56211700e+01, 9.88732834e+01, ..., 4.40814838e-05, 2.27930286e-05, 9.64050832e-06], [9.38575058e+01, 9.60387344e+01, 9.95178833e+01, ..., 4.01654543e-05, 1.81505584e-05, 8.73908903e-06]], dtype=float32) - RHI(time_lat_lon, lev)float3287.35 89.46 ... 1.815e-05 8.739e-06
- mdims :
- 9
- units :
- percent
- long_name :
- Relative humidity with respect to ice
- basename :
- RHI
array([[8.73468475e+01, 8.94625854e+01, 9.31331100e+01, ..., 3.14275967e-05, 1.22271085e-05, 8.82900440e-06], [8.66067123e+01, 8.86868210e+01, 9.22966232e+01, ..., 2.98583691e-05, 1.04015999e-05, 8.43694488e-06], [8.56140823e+01, 8.76887665e+01, 9.12698364e+01, ..., 2.96251437e-05, 1.07044179e-05, 9.32243165e-06], ..., [9.25081711e+01, 9.43432846e+01, 9.71473083e+01, ..., 4.38797069e-05, 2.67102241e-05, 9.65839445e-06], [9.35536499e+01, 9.56211700e+01, 9.88732834e+01, ..., 4.40814838e-05, 2.27930286e-05, 9.64050832e-06], [9.38575058e+01, 9.60387344e+01, 9.95178833e+01, ..., 4.01654543e-05, 1.81505584e-05, 8.73908903e-06]], dtype=float32) - RHW(time_lat_lon, lev)float3282.26 83.94 ... 0.0002166 0.0002172
- mdims :
- 9
- units :
- percent
- long_name :
- Relative humidity with respect to liquid
- basename :
- RHW
array([[8.2255363e+01, 8.3940842e+01, 8.6876579e+01, ..., 2.1726268e-04, 2.1540654e-04, 2.1281077e-04], [8.1629616e+01, 8.3288261e+01, 8.6178467e+01, ..., 2.1690367e-04, 2.1528378e-04, 2.1399544e-04], [8.0763603e+01, 8.2425003e+01, 8.5300423e+01, ..., 2.1676825e-04, 2.1551231e-04, 2.1503725e-04], ..., [8.6415276e+01, 8.7862564e+01, 9.0073730e+01, ..., 2.1620565e-04, 2.1655482e-04, 2.1797139e-04], [8.7961464e+01, 8.9617226e+01, 9.2217079e+01, ..., 2.1608094e-04, 2.1642998e-04, 2.1773181e-04], [8.8939125e+01, 9.0708588e+01, 9.3525269e+01, ..., 2.1627167e-04, 2.1664714e-04, 2.1724954e-04]], dtype=float32) - SNOWQM(time_lat_lon, lev)float322.413e-09 2.209e-09 ... 2.229e-24
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Grid box averaged snow amount
- basename :
- SNOWQM
array([[2.4134366e-09, 2.2085680e-09, 1.8306943e-09, ..., 2.5175845e-24, 2.2932725e-24, 2.2278446e-24], [2.0164521e-09, 1.8323438e-09, 1.4693483e-09, ..., 2.4975058e-24, 2.3096059e-24, 2.2531705e-24], [1.6153280e-09, 1.4304888e-09, 1.0998460e-09, ..., 2.5038842e-24, 2.3226227e-24, 2.2639099e-24], ..., [4.6601373e-05, 4.6617162e-05, 4.6587036e-05, ..., 2.4576474e-24, 2.3109761e-24, 2.1949836e-24], [5.4352276e-05, 5.4327065e-05, 5.4217737e-05, ..., 2.4732539e-24, 2.3043828e-24, 2.2061412e-24], [5.9973376e-05, 6.0003069e-05, 6.0095492e-05, ..., 2.4755895e-24, 2.2795694e-24, 2.2287567e-24]], dtype=float32) - SNOW_ICLD_VISTAU(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- Sampling_Sequence :
- rad_lwsw
- units :
- 1
- long_name :
- Snow in-cloud extinction visible sw optical depth
- basename :
- SNOW_ICLD_VISTAU
array([[0. , 0. , 0. , ..., 0. , 0. , 0. ], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [0.02150888, 0.04656317, 0.06306311, ..., 0. , 0. , 0. ], [0.02661125, 0.05730582, 0.07661083, ..., 0. , 0. , 0. ], [0.02438943, 0.05238835, 0.0696812 , ..., 0. , 0. , 0. ]], dtype=float32) - SOLIN(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Solar insolation
- basename :
- SOLIN
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - SWCF(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Shortwave cloud forcing
- basename :
- SWCF
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - T(time_lat_lon, lev)float32267.0 266.6 266.0 ... 261.2 265.1
- mdims :
- 9
- units :
- K
- long_name :
- Temperature
- basename :
- T
array([[266.9897 , 266.61496, 266.0163 , ..., 259.49014, 265.60776, 264.73633], [267.0793 , 266.70782, 266.1142 , ..., 260.034 , 267.46976, 265.3187 ], [267.16782, 266.80008, 266.2113 , ..., 260.1134 , 267.14853, 264.236 ], ..., [266.16086, 265.84958, 265.39474, ..., 255.83473, 256.99286, 263.9881 ], [266.82767, 266.4981 , 266.00067, ..., 255.77979, 258.70105, 263.99664], [267.6298 , 267.29352, 266.77948, ..., 256.78268, 261.2148 , 265.0888 ]], dtype=float32) - TGCLDCWP(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- units :
- kg/m2
- long_name :
- Total grid-box cloud water path (liquid and ice)
- basename :
- TGCLDCWP
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - TGCLDIWP(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- units :
- kg/m2
- long_name :
- Total grid-box cloud ice water path
- basename :
- TGCLDIWP
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - TGCLDLWP(time_lat_lon)float32dask.array<chunksize=(72,), meta=np.ndarray>
- units :
- kg/m2
- long_name :
- Total grid-box cloud liquid water path
- basename :
- TGCLDLWP
Array Chunk Bytes 288 B 288 B Shape (72,) (72,) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - TOT_CLD_VISTAU(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- Sampling_Sequence :
- rad_lwsw
- units :
- 1
- long_name :
- Total gbx cloud extinction visible sw optical depth
- basename :
- TOT_CLD_VISTAU
array([[0. , 0. , 0. , ..., 0. , 0. , 0. ], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [0.00537722, 0.01164079, 0.01576578, ..., 0. , 0. , 0. ], [0.00665281, 0.01432646, 0.01915271, ..., 0. , 0. , 0. ], [0.00609736, 0.01309709, 0.0174203 , ..., 0. , 0. , 0. ]], dtype=float32) - TOT_ICLD_VISTAU(time_lat_lon, lev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- Sampling_Sequence :
- rad_lwsw
- units :
- 1
- long_name :
- Total in-cloud extinction visible sw optical depth
- basename :
- TOT_ICLD_VISTAU
array([[0. , 0. , 0. , ..., 0. , 0. , 0. ], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [0.02150888, 0.04656317, 0.06306311, ..., 0. , 0. , 0. ], [0.02661125, 0.05730582, 0.07661083, ..., 0. , 0. , 0. ], [0.02438943, 0.05238835, 0.0696812 , ..., 0. , 0. , 0. ]], dtype=float32) - U(time_lat_lon, lev)float320.2622 0.2732 ... 91.51 105.0
- mdims :
- 9
- units :
- m/s
- long_name :
- Zonal wind
- basename :
- U
array([[ 0.26222718, 0.2731568 , 0.2674085 , ..., 50.331787 , 61.789238 , 58.816326 ], [ 0.533694 , 0.57024926, 0.5827104 , ..., 51.97362 , 68.08576 , 58.699936 ], [ 0.8422995 , 0.9089949 , 0.9417052 , ..., 54.015667 , 71.07826 , 56.519646 ], ..., [ 7.176378 , 9.861273 , 12.122251 , ..., 55.71363 , 70.53707 , 89.53816 ], [ 7.900169 , 10.590745 , 12.767541 , ..., 59.77514 , 80.72163 , 97.05116 ], [ 8.525111 , 11.240928 , 13.364264 , ..., 66.31535 , 91.51439 , 104.99406 ]], dtype=float32) - V(time_lat_lon, lev)float32-4.542 -4.865 -4.986 ... 13.4 17.55
- mdims :
- 9
- units :
- m/s
- long_name :
- Meridional wind
- basename :
- V
array([[-4.542304 , -4.8645105, -4.9855566, ..., 8.1667385, 17.39923 , -4.427489 ], [-3.9623654, -4.2424593, -4.339403 , ..., 9.337824 , 15.605526 , -3.1699805], [-3.2448413, -3.4740312, -3.54404 , ..., 8.085328 , 9.641141 , -2.3639543], ..., [ 6.970086 , 9.313141 , 10.691774 , ..., -1.9484863, 5.452907 , 5.499318 ], [ 6.7964725, 8.894286 , 10.129004 , ..., 3.9297893, 10.788858 , 12.646693 ], [ 6.369951 , 8.215149 , 9.270718 , ..., 8.591337 , 13.399579 , 17.549086 ]], dtype=float32) - WSUB(time_lat_lon, lev)float320.2996 0.4053 0.4906 ... 0.2 0.2
- mdims :
- 9
- units :
- m/s
- long_name :
- Diagnostic sub-grid vertical velocity
- basename :
- WSUB
array([[0.2996484 , 0.40532094, 0.49058613, ..., 0.2 , 0.2 , 0.2 ], [0.27994096, 0.3851581 , 0.46935824, ..., 0.2 , 0.2 , 0.2 ], [0.2582274 , 0.36366802, 0.44693393, ..., 0.2 , 0.2 , 0.2 ], ..., [0.6325026 , 0.5945988 , 0.48333636, ..., 0.2 , 0.2 , 0.2 ], [0.67820954, 0.64544046, 0.5395676 , ..., 0.2 , 0.2 , 0.2 ], [0.70120794, 0.67150956, 0.5715366 , ..., 0.2 , 0.2 , 0.2 ]], dtype=float32) - WSUBI(time_lat_lon, lev)float320.2996 0.4053 ... 0.001 0.001
- mdims :
- 9
- units :
- m/s
- long_name :
- Diagnostic sub-grid vertical velocity for ice
- basename :
- WSUBI
array([[0.2996484 , 0.40532094, 0.49058613, ..., 0.001 , 0.001 , 0.001 ], [0.27994096, 0.3851581 , 0.46935824, ..., 0.001 , 0.001 , 0.001 ], [0.2582274 , 0.36366802, 0.44693393, ..., 0.001 , 0.001 , 0.001 ], ..., [0.6325026 , 0.5945988 , 0.48333636, ..., 0.001 , 0.001 , 0.001 ], [0.67820954, 0.64544046, 0.5395676 , ..., 0.001 , 0.001 , 0.001 ], [0.70120794, 0.67150956, 0.5715366 , ..., 0.001 , 0.001 , 0.001 ]], dtype=float32) - Z3(time_lat_lon, lev)float3211.83 49.05 ... 5.794e+04 6.09e+04
- mdims :
- 9
- units :
- m
- long_name :
- Geopotential Height (above sea level)
- basename :
- Z3
array([[1.1827231e+01, 4.9051311e+01, 1.0870443e+02, ..., 5.5112230e+04, 5.8069688e+04, 6.1056691e+04], [1.1831173e+01, 4.9068027e+01, 1.0874337e+02, ..., 5.5076398e+04, 5.8047406e+04, 6.1048176e+04], [1.1835021e+01, 4.9084423e+01, 1.0878167e+02, ..., 5.5042754e+04, 5.8012398e+04, 6.1005262e+04], ..., [8.9983124e+01, 1.2709759e+02, 1.8655995e+02, ..., 5.4989383e+04, 5.7877730e+04, 6.0812000e+04], [9.0013542e+01, 1.2722248e+02, 1.8682597e+02, ..., 5.5006480e+04, 5.7904141e+04, 6.0848078e+04], [9.0050064e+01, 1.2737388e+02, 1.8715572e+02, ..., 5.5017727e+04, 5.7935191e+04, 6.0899438e+04]], dtype=float32) - bc_a1(time_lat_lon, lev)float321.782e-12 1.784e-12 ... 4.415e-22
- mdims :
- 9
- units :
- kg/kg
- long_name :
- bc_a1 concentration
- basename :
- bc_a1
array([[1.7816114e-12, 1.7840002e-12, 1.7883274e-12, ..., 1.4724844e-20, 1.7302145e-21, 1.1319374e-21], [1.8329763e-12, 1.8347112e-12, 1.8378751e-12, ..., 1.4708845e-20, 1.5664211e-21, 1.1887907e-21], [1.9403762e-12, 1.9419160e-12, 1.9446990e-12, ..., 1.5933869e-20, 1.6690077e-21, 1.3432827e-21], ..., [3.5497592e-12, 3.5417795e-12, 3.5164989e-12, ..., 9.2568829e-21, 1.1495995e-21, 4.2357307e-22], [3.1481714e-12, 3.1395912e-12, 3.1133486e-12, ..., 1.1432430e-20, 1.1096547e-21, 4.2809813e-22], [2.5720678e-12, 2.5574321e-12, 2.5137555e-12, ..., 1.2027146e-20, 9.9041933e-22, 4.4145166e-22]], dtype=float32) - bc_a3(time_lat_lon, lev)float327.966e-14 8.098e-14 ... 1.005e-36
- mdims :
- 9
- units :
- kg/kg
- long_name :
- bc_a3 concentration
- basename :
- bc_a3
array([[7.96646413e-14, 8.09841289e-14, 8.66015904e-14, ..., 1.00000004e-36, 1.00050558e-36, 1.00000004e-36], [7.78791975e-14, 7.89440805e-14, 8.38406222e-14, ..., 1.00003950e-36, 1.00019653e-36, 1.00000004e-36], [1.82066981e-13, 1.94093588e-13, 1.93940770e-13, ..., 1.00000748e-36, 1.00021205e-36, 1.00369516e-36], ..., [1.06857767e-13, 1.06577988e-13, 1.05717715e-13, ..., 1.00000004e-36, 1.00000004e-36, 1.00878010e-36], [8.50353723e-14, 8.47215297e-14, 8.38113216e-14, ..., 1.00000004e-36, 1.00000004e-36, 1.00836855e-36], [6.37070353e-14, 6.30605594e-14, 6.13599205e-14, ..., 1.00000004e-36, 1.00000004e-36, 1.00483189e-36]], dtype=float32) - bc_a4(time_lat_lon, lev)float322.008e-12 2.01e-12 ... 1.327e-18
- mdims :
- 9
- units :
- kg/kg
- long_name :
- bc_a4 concentration
- basename :
- bc_a4
array([[2.0082790e-12, 2.0099554e-12, 2.0134575e-12, ..., 2.2444537e-18, 1.7253130e-18, 1.6471388e-18], [2.0631329e-12, 2.0646002e-12, 2.0676362e-12, ..., 2.2291471e-18, 1.8015125e-18, 1.6567445e-18], [2.1179243e-12, 2.1191419e-12, 2.1216546e-12, ..., 2.3124961e-18, 1.8370294e-18, 1.6014339e-18], ..., [8.4393898e-12, 8.3610566e-12, 8.1214124e-12, ..., 1.8220442e-18, 1.7020915e-18, 1.4774114e-18], [7.2563140e-12, 7.2064403e-12, 7.0588396e-12, ..., 1.8731581e-18, 1.6343458e-18, 1.4112126e-18], [6.1419407e-12, 6.0789390e-12, 5.8944707e-12, ..., 1.9198819e-18, 1.5715146e-18, 1.3274013e-18]], dtype=float32) - bc_c1(time_lat_lon, lev)float320.0 0.0 0.0 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- bc_c1 in cloud water
- basename :
- bc_c1
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - bc_c3(time_lat_lon, lev)float320.0 0.0 0.0 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- bc_c3 in cloud water
- basename :
- bc_c3
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - bc_c4(time_lat_lon, lev)float320.0 0.0 0.0 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- bc_c4 in cloud water
- basename :
- bc_c4
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - dei_cloud(time_lat_lon, lev)float3250.0 50.0 300.2 ... 27.26 27.26
- mdims :
- 9
- units :
- micrometers
- long_name :
- ice radiative effective diameter in cloud
- basename :
- dei_cloud
array([[ 50. , 50. , 300.1505 , ..., 27.262814, 27.262814, 27.262814], [ 50. , 50. , 167.78566 , ..., 27.262814, 27.262814, 27.262814], [ 50. , 50. , 50. , ..., 27.262814, 27.262814, 27.262814], ..., [ 50. , 50. , 50. , ..., 27.262814, 27.262814, 27.262814], [ 50. , 50. , 50. , ..., 27.262814, 27.262814, 27.262814], [ 50. , 50. , 50. , ..., 27.262814, 27.262814, 27.262814]], dtype=float32) - dgnd_a01(time_lat_lon, lev)float321.538e-07 1.538e-07 ... 5.35e-08
- mdims :
- 9
- units :
- m
- long_name :
- dry dgnum, interstitial, mode 01
- basename :
- dgnd_a01
array([[1.5383517e-07, 1.5380071e-07, 1.5373723e-07, ..., 5.3499999e-08, 5.3499999e-08, 5.3499999e-08], [1.5361731e-07, 1.5357874e-07, 1.5350874e-07, ..., 5.3499999e-08, 5.3499999e-08, 5.3499999e-08], [1.5334609e-07, 1.5330185e-07, 1.5322331e-07, ..., 5.3499999e-08, 5.3499999e-08, 5.3499999e-08], ..., [1.4555611e-07, 1.4550541e-07, 1.4534918e-07, ..., 5.3499999e-08, 5.3499999e-08, 5.3499999e-08], [1.4617697e-07, 1.4612306e-07, 1.4596088e-07, ..., 5.3499999e-08, 5.3499999e-08, 5.3499999e-08], [1.4563255e-07, 1.4553440e-07, 1.4524052e-07, ..., 5.3499999e-08, 5.3499999e-08, 5.3499999e-08]], dtype=float32) - dgnd_a02(time_lat_lon, lev)float325.079e-08 5.078e-08 ... 1.433e-08
- mdims :
- 9
- units :
- m
- long_name :
- dry dgnum, interstitial, mode 02
- basename :
- dgnd_a02
array([[5.0785992e-08, 5.0783438e-08, 5.0777771e-08, ..., 1.5418745e-08, 1.4904858e-08, 1.4804867e-08], [5.0906017e-08, 5.0897835e-08, 5.0882083e-08, ..., 1.5427199e-08, 1.5000072e-08, 1.4814503e-08], [5.1248978e-08, 5.1235929e-08, 5.1211661e-08, ..., 1.5511311e-08, 1.5030167e-08, 1.4749315e-08], ..., [5.1237581e-08, 5.1225669e-08, 5.1187705e-08, ..., 1.5056738e-08, 1.4856580e-08, 1.4496997e-08], [5.1001450e-08, 5.0973028e-08, 5.0887341e-08, ..., 1.5061723e-08, 1.4783034e-08, 1.4421539e-08], [4.9726740e-08, 4.9658336e-08, 4.9453444e-08, ..., 1.5084739e-08, 1.4703829e-08, 1.4329004e-08]], dtype=float32) - dgnd_a03(time_lat_lon, lev)float321.575e-06 1.574e-06 ... 1e-06 1e-06
- mdims :
- 9
- units :
- m
- long_name :
- dry dgnum, interstitial, mode 03
- basename :
- dgnd_a03
array([[1.5748991e-06, 1.5739555e-06, 1.5722284e-06, ..., 1.0000000e-06, 1.0000000e-06, 1.0000000e-06], [1.5579387e-06, 1.5570454e-06, 1.5554376e-06, ..., 1.0000000e-06, 1.0000000e-06, 1.0000000e-06], [1.5515798e-06, 1.5503101e-06, 1.5480921e-06, ..., 1.0000000e-06, 1.0000000e-06, 1.0000000e-06], ..., [1.4041104e-06, 1.4050420e-06, 1.4078915e-06, ..., 1.0000000e-06, 1.0000000e-06, 1.0000000e-06], [1.3612458e-06, 1.3619272e-06, 1.3639561e-06, ..., 1.0000000e-06, 1.0000000e-06, 1.0000000e-06], [1.3317351e-06, 1.3319673e-06, 1.3326531e-06, ..., 1.0000000e-06, 1.0000000e-06, 1.0000000e-06]], dtype=float32) - dgnd_a04(time_lat_lon, lev)float321e-07 1e-07 ... 2.692e-08 2.619e-08
- mdims :
- 9
- units :
- m
- long_name :
- dry dgnum, interstitial, mode 04
- basename :
- dgnd_a04
array([[1.0000000e-07, 1.0000000e-07, 1.0000000e-07, ..., 2.8425202e-08, 2.7221509e-08, 2.6974408e-08], [1.0000000e-07, 1.0000000e-07, 1.0000000e-07, ..., 2.8401072e-08, 2.7358036e-08, 2.6987667e-08], [1.0000000e-07, 1.0000000e-07, 1.0000000e-07, ..., 2.8488294e-08, 2.7429099e-08, 2.6916378e-08], ..., [1.0000000e-07, 1.0000000e-07, 1.0000000e-07, ..., 2.7756624e-08, 2.7242509e-08, 2.6530563e-08], [1.0000000e-07, 1.0000000e-07, 1.0000000e-07, ..., 2.7855181e-08, 2.7089474e-08, 2.6377634e-08], [1.0000000e-07, 1.0000000e-07, 1.0000000e-07, ..., 2.7936485e-08, 2.6917320e-08, 2.6192012e-08]], dtype=float32) - dgnw_a01(time_lat_lon, lev)float322.199e-07 2.251e-07 ... 5.35e-08
- mdims :
- 9
- units :
- m
- long_name :
- wet dgnum, interstitial, mode 01
- basename :
- dgnw_a01
array([[2.1989361e-07, 2.2511547e-07, 2.3644169e-07, ..., 5.3499999e-08, 5.3499999e-08, 5.3499999e-08], [2.1691892e-07, 2.2172183e-07, 2.3203027e-07, ..., 5.3499999e-08, 5.3499999e-08, 5.3499999e-08], [2.1316407e-07, 2.1756902e-07, 2.2687054e-07, ..., 5.3499999e-08, 5.3499999e-08, 5.3499999e-08], ..., [2.1243163e-07, 2.1806123e-07, 2.2862820e-07, ..., 5.3499999e-08, 5.3499999e-08, 5.3499999e-08], [2.2180228e-07, 2.2993616e-07, 2.4709547e-07, ..., 5.3499999e-08, 5.3499999e-08, 5.3499999e-08], [2.2818399e-07, 2.3833068e-07, 2.6144639e-07, ..., 5.3499999e-08, 5.3499999e-08, 5.3499999e-08]], dtype=float32) - dgnw_a02(time_lat_lon, lev)float327.364e-08 7.532e-08 ... 1.433e-08
- mdims :
- 9
- units :
- m
- long_name :
- wet dgnum, interstitial, mode 02
- basename :
- dgnw_a02
array([[7.3642909e-08, 7.5323349e-08, 7.8909913e-08, ..., 1.5418745e-08, 1.4904858e-08, 1.4804867e-08], [7.2982544e-08, 7.4538299e-08, 7.7827472e-08, ..., 1.5427199e-08, 1.5000072e-08, 1.4814503e-08], [7.2370959e-08, 7.3816352e-08, 7.6825486e-08, ..., 1.5511311e-08, 1.5030167e-08, 1.4749315e-08], ..., [7.5033796e-08, 7.6863103e-08, 8.0225057e-08, ..., 1.5056738e-08, 1.4856580e-08, 1.4496997e-08], [7.7376662e-08, 7.9930487e-08, 8.5144556e-08, ..., 1.5061723e-08, 1.4783034e-08, 1.4421539e-08], [7.7635661e-08, 8.0660854e-08, 8.7229083e-08, ..., 1.5084739e-08, 1.4703829e-08, 1.4329004e-08]], dtype=float32) - dgnw_a03(time_lat_lon, lev)float322.984e-06 3.078e-06 ... 1e-06 1e-06
- mdims :
- 9
- units :
- m
- long_name :
- wet dgnum, interstitial, mode 03
- basename :
- dgnw_a03
array([[2.9843213e-06, 3.0775045e-06, 3.2767675e-06, ..., 1.0000000e-06, 1.0000000e-06, 1.0000000e-06], [2.9187961e-06, 3.0049403e-06, 3.1871832e-06, ..., 1.0000000e-06, 1.0000000e-06, 1.0000000e-06], [2.8585362e-06, 2.9380710e-06, 3.1036714e-06, ..., 1.0000000e-06, 1.0000000e-06, 1.0000000e-06], ..., [2.8604568e-06, 2.9661530e-06, 3.1684508e-06, ..., 1.0000000e-06, 1.0000000e-06, 1.0000000e-06], [2.8831969e-06, 3.0236331e-06, 3.3209581e-06, ..., 1.0000000e-06, 1.0000000e-06, 1.0000000e-06], [2.9022601e-06, 3.0689666e-06, 3.4490818e-06, ..., 1.0000000e-06, 1.0000000e-06, 1.0000000e-06]], dtype=float32) - dgnw_a04(time_lat_lon, lev)float321e-07 1e-07 ... 2.692e-08 2.619e-08
- mdims :
- 9
- units :
- m
- long_name :
- wet dgnum, interstitial, mode 04
- basename :
- dgnw_a04
array([[1.00000172e-07, 1.00000186e-07, 1.00000229e-07, ..., 2.84252017e-08, 2.72215086e-08, 2.69744085e-08], [1.00000157e-07, 1.00000179e-07, 1.00000214e-07, ..., 2.84010717e-08, 2.73580358e-08, 2.69876672e-08], [1.00000150e-07, 1.00000165e-07, 1.00000193e-07, ..., 2.84882944e-08, 2.74290990e-08, 2.69163785e-08], ..., [1.00000108e-07, 1.00000122e-07, 1.00000150e-07, ..., 2.77566237e-08, 2.72425087e-08, 2.65305626e-08], [1.00000150e-07, 1.00000179e-07, 1.00000236e-07, ..., 2.78551813e-08, 2.70894738e-08, 2.63776343e-08], [1.00000200e-07, 1.00000243e-07, 1.00000342e-07, ..., 2.79364851e-08, 2.69173199e-08, 2.61920121e-08]], dtype=float32) - dst_a1(time_lat_lon, lev)float324.228e-12 4.232e-12 ... 2.129e-21
- mdims :
- 9
- units :
- kg/kg
- long_name :
- dst_a1 concentration
- basename :
- dst_a1
array([[4.22774359e-12, 4.23225907e-12, 4.24103591e-12, ..., 6.44066542e-20, 8.46019893e-21, 5.80486354e-21], [4.22442246e-12, 4.22720929e-12, 4.23279510e-12, ..., 6.47261915e-20, 7.81291637e-21, 5.93339279e-21], [4.41989457e-12, 4.42209810e-12, 4.42650560e-12, ..., 7.03539988e-20, 8.07361801e-21, 6.38818258e-21], ..., [1.14393920e-11, 1.14360041e-11, 1.14228930e-11, ..., 4.03745629e-20, 5.46040744e-21, 2.13495296e-21], [1.03578509e-11, 1.03396736e-11, 1.02846646e-11, ..., 4.92937090e-20, 5.16759116e-21, 2.11216066e-21], [8.65910035e-12, 8.62471119e-12, 8.52298353e-12, ..., 5.16436677e-20, 4.62160470e-21, 2.12915462e-21]], dtype=float32) - dst_a3(time_lat_lon, lev)float329.09e-12 9.112e-12 ... 1.005e-36
- mdims :
- 9
- units :
- kg/kg
- long_name :
- dst_a3 concentration
- basename :
- dst_a3
array([[9.09042459e-12, 9.11186317e-12, 9.17691357e-12, ..., 1.00000004e-36, 1.00050558e-36, 1.00000004e-36], [8.63755768e-12, 8.64679335e-12, 8.68594953e-12, ..., 1.00003950e-36, 1.00019653e-36, 1.00000004e-36], [9.81701265e-12, 9.88991094e-12, 9.87373464e-12, ..., 1.00000748e-36, 1.00021205e-36, 1.00369516e-36], ..., [5.86975885e-11, 5.86648993e-11, 5.85633139e-11, ..., 1.00000004e-36, 1.00000004e-36, 1.00878010e-36], [5.08222527e-11, 5.07240361e-11, 5.04403255e-11, ..., 1.00000004e-36, 1.00000004e-36, 1.00836855e-36], [4.05322789e-11, 4.02749917e-11, 3.95399685e-11, ..., 1.00000004e-36, 1.00000004e-36, 1.00483189e-36]], dtype=float32) - dst_c1(time_lat_lon, lev)float320.0 0.0 0.0 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- dst_c1 in cloud water
- basename :
- dst_c1
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - dst_c3(time_lat_lon, lev)float320.0 0.0 0.0 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- dst_c3 in cloud water
- basename :
- dst_c3
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - lambda_cloud(time_lat_lon, lev)float322.52e+05 2.52e+05 ... 0.0 0.0
- mdims :
- 9
- units :
- 1/meter
- long_name :
- lambda in cloud
- basename :
- lambda_cloud
array([[252000. , 252000. , 271525.3, ..., 0. , 0. , 0. ], [252000. , 252000. , 0. , ..., 0. , 0. , 0. ], [252000. , 252000. , 252000. , ..., 0. , 0. , 0. ], ..., [252000. , 252000. , 252000. , ..., 0. , 0. , 0. ], [252000. , 252000. , 252000. , ..., 0. , 0. , 0. ], [252000. , 252000. , 252000. , ..., 0. , 0. , 0. ]], dtype=float32) - mom_a1(time_lat_lon, lev)float326.74e-12 6.72e-12 ... 1.201e-26
- mdims :
- 9
- units :
- kg/kg
- long_name :
- mom_a1 concentration
- basename :
- mom_a1
array([[6.73974244e-12, 6.72033653e-12, 6.68515807e-12, ..., 5.43284315e-25, 6.18996288e-26, 4.13196529e-26], [6.23085090e-12, 6.21409911e-12, 6.18423151e-12, ..., 5.51787003e-25, 5.51686546e-26, 4.18887698e-26], [5.83012891e-12, 5.81656858e-12, 5.79282802e-12, ..., 6.07118347e-25, 5.77081150e-26, 4.60885574e-26], ..., [3.41564034e-11, 3.42837841e-11, 3.46313186e-11, ..., 3.37366252e-25, 3.47745541e-26, 1.16681472e-26], [3.61434840e-11, 3.60908074e-11, 3.59184280e-11, ..., 4.15161421e-25, 3.32795425e-26, 1.16314105e-26], [3.66858800e-11, 3.65697542e-11, 3.62231044e-11, ..., 4.33683925e-25, 2.97512635e-26, 1.20109512e-26]], dtype=float32) - mom_a2(time_lat_lon, lev)float323.87e-14 3.829e-14 ... 8.16e-21
- mdims :
- 9
- units :
- kg/kg
- long_name :
- mom_a2 concentration
- basename :
- mom_a2
array([[3.86956905e-14, 3.82888166e-14, 3.75505596e-14, ..., 1.30988808e-20, 1.04955883e-20, 1.01006201e-20], [3.07236909e-14, 3.03936631e-14, 2.98049617e-14, ..., 1.30307079e-20, 1.09365467e-20, 1.01552180e-20], [2.33709536e-14, 2.31210975e-14, 2.26865510e-14, ..., 1.34793330e-20, 1.10908110e-20, 9.79285718e-21], ..., [2.04539429e-13, 2.05499449e-13, 2.08283531e-13, ..., 1.06412480e-20, 1.02129300e-20, 9.06547381e-21], [1.75003729e-13, 1.74836016e-13, 1.74329531e-13, ..., 1.08946626e-20, 9.86122236e-21, 8.67260825e-21], [1.35161833e-13, 1.34174423e-13, 1.31293834e-13, ..., 1.11158203e-20, 9.51978499e-21, 8.16048086e-21]], dtype=float32) - mom_a3(time_lat_lon, lev)float322.536e-13 2.586e-13 ... 1.005e-36
- mdims :
- 9
- units :
- kg/kg
- long_name :
- mom_a3 concentration
- basename :
- mom_a3
array([[2.53627441e-13, 2.58551806e-13, 2.79789267e-13, ..., 1.00000004e-36, 1.00050558e-36, 1.00000004e-36], [2.47831703e-13, 2.51278165e-13, 2.67773867e-13, ..., 1.00003950e-36, 1.00019653e-36, 1.00000004e-36], [5.84717875e-13, 6.20913368e-13, 6.19221905e-13, ..., 1.00000748e-36, 1.00021205e-36, 1.00369516e-36], ..., [6.93943275e-13, 6.92445396e-13, 6.87711119e-13, ..., 1.00000004e-36, 1.00000004e-36, 1.00878010e-36], [6.08017217e-13, 6.05474492e-13, 5.98078661e-13, ..., 1.00000004e-36, 1.00000004e-36, 1.00836855e-36], [5.13321211e-13, 5.08033773e-13, 4.94894436e-13, ..., 1.00000004e-36, 1.00000004e-36, 1.00483189e-36]], dtype=float32) - mom_a4(time_lat_lon, lev)float325.877e-17 5.883e-17 ... 3.232e-23
- mdims :
- 9
- units :
- kg/kg
- long_name :
- mom_a4 concentration
- basename :
- mom_a4
array([[5.87696671e-17, 5.88308322e-17, 5.89544394e-17, ..., 5.42586293e-23, 4.19554536e-23, 4.00615280e-23], [5.91907021e-17, 5.92515759e-17, 5.93707958e-17, ..., 5.39316496e-23, 4.38300364e-23, 4.03459344e-23], [5.91552987e-17, 5.92185416e-17, 5.93376160e-17, ..., 5.60275647e-23, 4.46994274e-23, 3.90130056e-23], ..., [1.08853706e-16, 1.09068145e-16, 1.09688728e-16, ..., 4.44418328e-23, 4.12792610e-23, 3.58116219e-23], [1.15966500e-16, 1.16128151e-16, 1.16619562e-16, ..., 4.55247937e-23, 3.97217719e-23, 3.42584905e-23], [1.21383448e-16, 1.21569914e-16, 1.22129234e-16, ..., 4.65010500e-23, 3.82451284e-23, 3.23208398e-23]], dtype=float32) - mom_c1(time_lat_lon, lev)float320.0 0.0 0.0 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- mom_c1 in cloud water
- basename :
- mom_c1
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - mom_c2(time_lat_lon, lev)float320.0 0.0 0.0 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- mom_c2 in cloud water
- basename :
- mom_c2
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - mom_c3(time_lat_lon, lev)float320.0 0.0 0.0 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- mom_c3 in cloud water
- basename :
- mom_c3
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - mom_c4(time_lat_lon, lev)float320.0 0.0 0.0 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- mom_c4 in cloud water
- basename :
- mom_c4
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - mu_cloud(time_lat_lon, lev)float325.3 5.3 12.58 12.58 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- 1
- long_name :
- mu in cloud
- basename :
- mu_cloud
array([[ 5.3 , 5.3 , 12.576265, ..., 0. , 0. , 0. ], [ 5.3 , 5.3 , 0. , ..., 0. , 0. , 0. ], [ 5.3 , 5.3 , 5.3 , ..., 0. , 0. , 0. ], ..., [ 5.3 , 5.3 , 5.3 , ..., 0. , 0. , 0. ], [ 5.3 , 5.3 , 5.3 , ..., 0. , 0. , 0. ], [ 5.3 , 5.3 , 5.3 , ..., 0. , 0. , 0. ]], dtype=float32) - ncl_a1(time_lat_lon, lev)float324.32e-11 4.313e-11 ... 1.155e-21
- mdims :
- 9
- units :
- kg/kg
- long_name :
- ncl_a1 concentration
- basename :
- ncl_a1
array([[4.3201026e-11, 4.3128036e-11, 4.2995614e-11, ..., 5.4001678e-20, 5.6926222e-21, 3.7042889e-21], [4.0537521e-11, 4.0470845e-11, 4.0351684e-11, ..., 5.4427328e-20, 5.0217570e-21, 3.8436058e-21], [3.8661137e-11, 3.8604182e-11, 3.8503943e-11, ..., 5.9934845e-20, 5.3985276e-21, 4.3588824e-21], ..., [5.8422787e-11, 5.8192829e-11, 5.7474647e-11, ..., 3.3680281e-20, 3.3850511e-21, 1.0971982e-21], [5.7764082e-11, 5.7466677e-11, 5.6582364e-11, ..., 4.1724764e-20, 3.2686042e-21, 1.1057821e-21], [5.4448421e-11, 5.4040127e-11, 5.2844451e-11, ..., 4.3768294e-20, 2.9090185e-21, 1.1546363e-21]], dtype=float32) - ncl_a2(time_lat_lon, lev)float321.85e-13 1.835e-13 ... 4.3e-16
- mdims :
- 9
- units :
- kg/kg
- long_name :
- ncl_a2 concentration
- basename :
- ncl_a2
array([[1.8502070e-13, 1.8351752e-13, 1.8078198e-13, ..., 6.5833831e-16, 5.4672710e-16, 5.2951402e-16], [1.4996630e-13, 1.4877214e-13, 1.4663298e-13, ..., 6.5689115e-16, 5.6943574e-16, 5.3167252e-16], [1.2019540e-13, 1.1931058e-13, 1.1776185e-13, ..., 6.7842109e-16, 5.7379552e-16, 5.1036424e-16], ..., [3.5704743e-13, 3.5463597e-13, 3.4743733e-13, ..., 5.3908035e-16, 5.2711369e-16, 4.7432347e-16], [3.2273208e-13, 3.2028490e-13, 3.1323872e-13, ..., 5.4700927e-16, 5.1059199e-16, 4.5534019e-16], [2.7516281e-13, 2.7167073e-13, 2.6163094e-13, ..., 5.5367013e-16, 4.9528626e-16, 4.3004882e-16]], dtype=float32) - ncl_a3(time_lat_lon, lev)float328.369e-10 8.35e-10 ... 1.005e-36
- mdims :
- 9
- units :
- kg/kg
- long_name :
- ncl_a3 concentration
- basename :
- ncl_a3
array([[8.36920033e-10, 8.35040925e-10, 8.33173364e-10, ..., 1.00000004e-36, 1.00050558e-36, 1.00000004e-36], [7.54990848e-10, 7.53272278e-10, 7.51473939e-10, ..., 1.00003950e-36, 1.00019653e-36, 1.00000004e-36], [7.26354754e-10, 7.28928584e-10, 7.25543237e-10, ..., 1.00000748e-36, 1.00021205e-36, 1.00369516e-36], ..., [8.71872852e-10, 8.69607664e-10, 8.62546090e-10, ..., 1.00000004e-36, 1.00000004e-36, 1.00878010e-36], [7.76307130e-10, 7.73043130e-10, 7.63481667e-10, ..., 1.00000004e-36, 1.00000004e-36, 1.00836855e-36], [6.78960832e-10, 6.73573641e-10, 6.58102628e-10, ..., 1.00000004e-36, 1.00000004e-36, 1.00483189e-36]], dtype=float32) - ncl_c1(time_lat_lon, lev)float320.0 0.0 0.0 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- ncl_c1 in cloud water
- basename :
- ncl_c1
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - ncl_c2(time_lat_lon, lev)float320.0 0.0 0.0 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- ncl_c2 in cloud water
- basename :
- ncl_c2
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - ncl_c3(time_lat_lon, lev)float320.0 0.0 0.0 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- ncl_c3 in cloud water
- basename :
- ncl_c3
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - num_a1(time_lat_lon, lev)float321.86e+07 1.861e+07 ... 0.04611
- mdims :
- 9
- units :
- 1/kg
- long_name :
- num_a1 concentration
- basename :
- num_a1
array([[1.8599182e+07, 1.8613988e+07, 1.8642532e+07, ..., 1.4345158e+00, 1.9578731e-01, 1.3382529e-01], [1.8742460e+07, 1.8753600e+07, 1.8775244e+07, ..., 1.4458982e+00, 1.8763921e-01, 1.4174139e-01], [1.9397822e+07, 1.9408690e+07, 1.9429084e+07, ..., 1.6012802e+00, 2.0409805e-01, 1.5415908e-01], ..., [3.2224592e+07, 3.2238866e+07, 3.2268732e+07, ..., 9.0460289e-01, 1.3279216e-01, 4.4114098e-02], [3.0782620e+07, 3.0738350e+07, 3.0600744e+07, ..., 1.1186939e+00, 1.2943783e-01, 4.4507414e-02], [2.8127992e+07, 2.8050288e+07, 2.7818734e+07, ..., 1.1764649e+00, 1.1681487e-01, 4.6113059e-02]], dtype=float32) - num_a2(time_lat_lon, lev)float321.002e+07 1.003e+07 ... 3.83e+06
- mdims :
- 9
- units :
- 1/kg
- long_name :
- num_a2 concentration
- basename :
- num_a2
array([[10024589. , 10027550. , 10034107. , ..., 4753257. , 4350748.5, 4295866. ], [10026496. , 10030834. , 10039808. , ..., 4740030.5, 4461686. , 4314747.5], [10175696. , 10181941. , 10194128. , ..., 4835031. , 4471276.5, 4192521. ], ..., [24253308. , 24225560. , 24137434. , ..., 4179986. , 4206137. , 4039863.5], [22555044. , 22522964. , 22427530. , ..., 4229333.5, 4134859.5, 3953772. ], [20903662. , 20844644. , 20672252. , ..., 4250392. , 4083001.2, 3829992.8]], dtype=float32) - num_a3(time_lat_lon, lev)float324.635e+04 4.629e+04 ... 1e-05 1e-05
- mdims :
- 9
- units :
- 1/kg
- long_name :
- num_a3 concentration
- basename :
- num_a3
array([[4.6353160e+04, 4.6287258e+04, 4.6167824e+04, ..., 9.9999997e-06, 9.9999997e-06, 9.9999997e-06], [4.3343113e+04, 4.3283758e+04, 4.3178164e+04, ..., 9.9999997e-06, 9.9999997e-06, 9.9999997e-06], [4.1218855e+04, 4.1168973e+04, 4.1082191e+04, ..., 9.9999997e-06, 9.9999997e-06, 9.9999997e-06], ..., [7.1719000e+04, 7.1397148e+04, 7.0408734e+04, ..., 9.9999997e-06, 9.9999997e-06, 9.9999997e-06], [6.9958312e+04, 6.9566820e+04, 6.8419844e+04, ..., 9.9999997e-06, 9.9999997e-06, 9.9999997e-06], [6.5092113e+04, 6.4546461e+04, 6.2973477e+04, ..., 9.9999997e-06, 9.9999997e-06, 9.9999997e-06]], dtype=float32) - num_a4(time_lat_lon, lev)float322.31e+06 2.313e+06 ... 1.078e+03
- mdims :
- 9
- units :
- 1/kg
- long_name :
- num_a4 concentration
- basename :
- num_a4
array([[2.3100808e+06, 2.3125705e+06, 2.3176360e+06, ..., 1.4148236e+03, 1.2448466e+03, 1.2205455e+03], [2.4000978e+06, 2.4023542e+06, 2.4068838e+06, ..., 1.4102493e+03, 1.2813918e+03, 1.2286875e+03], [2.4777172e+06, 2.4796962e+06, 2.4836152e+06, ..., 1.4527782e+03, 1.2979293e+03, 1.1986926e+03], ..., [7.7126955e+06, 7.6487645e+06, 7.4526140e+06, ..., 1.2511407e+03, 1.2213602e+03, 1.1446022e+03], [6.6087465e+06, 6.5688270e+06, 6.4504570e+06, ..., 1.2664630e+03, 1.1964390e+03, 1.1154211e+03], [5.5987820e+06, 5.5479810e+06, 5.3991150e+06, ..., 1.2804136e+03, 1.1748929e+03, 1.0777002e+03]], dtype=float32) - num_c1(time_lat_lon, lev)float320.0 0.0 0.0 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- 1/kg
- long_name :
- num_c1 in cloud water
- basename :
- num_c1
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - num_c2(time_lat_lon, lev)float320.0 0.0 0.0 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- 1/kg
- long_name :
- num_c2 in cloud water
- basename :
- num_c2
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - num_c3(time_lat_lon, lev)float320.0 0.0 0.0 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- 1/kg
- long_name :
- num_c3 in cloud water
- basename :
- num_c3
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - num_c4(time_lat_lon, lev)float320.0 0.0 0.0 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- 1/kg
- long_name :
- num_c4 in cloud water
- basename :
- num_c4
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - pom_a1(time_lat_lon, lev)float322.438e-12 2.445e-12 ... 2.49e-21
- mdims :
- 9
- units :
- kg/kg
- long_name :
- pom_a1 concentration
- basename :
- pom_a1
array([[2.4381541e-12, 2.4450580e-12, 2.4577538e-12, ..., 8.3421236e-20, 9.8092804e-21, 6.4215077e-21], [2.4367431e-12, 2.4434075e-12, 2.4555690e-12, ..., 8.3330556e-20, 8.8798881e-21, 6.7386551e-21], [2.5618763e-12, 2.5686161e-12, 2.5806579e-12, ..., 9.0266595e-20, 9.4600147e-21, 7.6141232e-21], ..., [9.0569080e-12, 9.0494166e-12, 9.0242241e-12, ..., 5.2587015e-20, 6.4938767e-21, 2.3959945e-21], [8.2421275e-12, 8.2249598e-12, 8.1727793e-12, ..., 6.4815340e-20, 6.2607152e-21, 2.4175725e-21], [6.8903052e-12, 6.8589752e-12, 6.7659645e-12, ..., 6.8054947e-20, 5.5862172e-21, 2.4899839e-21]], dtype=float32) - pom_a3(time_lat_lon, lev)float321.1e-13 1.12e-13 ... 1.005e-36
- mdims :
- 9
- units :
- kg/kg
- long_name :
- pom_a3 concentration
- basename :
- pom_a3
array([[1.10009698e-13, 1.11981130e-13, 1.20121429e-13, ..., 1.00000004e-36, 1.00050558e-36, 1.00000004e-36], [1.02022103e-13, 1.03590632e-13, 1.10508845e-13, ..., 1.00003950e-36, 1.00019653e-36, 1.00000004e-36], [2.43686283e-13, 2.60037760e-13, 2.59908685e-13, ..., 1.00000748e-36, 1.00021205e-36, 1.00369516e-36], ..., [3.36563568e-13, 3.35828858e-13, 3.33563960e-13, ..., 1.00000004e-36, 1.00000004e-36, 1.00878010e-36], [2.76576179e-13, 2.75625307e-13, 2.72870539e-13, ..., 1.00000004e-36, 1.00000004e-36, 1.00836855e-36], [2.12604443e-13, 2.10560898e-13, 2.05185614e-13, ..., 1.00000004e-36, 1.00000004e-36, 1.00483189e-36]], dtype=float32) - pom_a4(time_lat_lon, lev)float322.182e-12 2.185e-12 ... 2.662e-17
- mdims :
- 9
- units :
- kg/kg
- long_name :
- pom_a4 concentration
- basename :
- pom_a4
array([[2.1824196e-12, 2.1846917e-12, 2.1893305e-12, ..., 4.4655438e-17, 3.4512870e-17, 3.2925279e-17], [2.2761643e-12, 2.2782414e-12, 2.2824212e-12, ..., 4.4399184e-17, 3.6064066e-17, 3.3196065e-17], [2.3483682e-12, 2.3502439e-12, 2.3539530e-12, ..., 4.6163825e-17, 3.6816069e-17, 3.2130951e-17], ..., [7.1697431e-12, 7.1079501e-12, 6.9189242e-12, ..., 3.6783058e-17, 3.3936967e-17, 2.9372859e-17], [6.2887560e-12, 6.2496995e-12, 6.1341427e-12, ..., 3.7626213e-17, 3.2690373e-17, 2.8134090e-17], [5.4620011e-12, 5.4132775e-12, 5.2706641e-12, ..., 3.8369196e-17, 3.1495296e-17, 2.6617248e-17]], dtype=float32) - pom_c1(time_lat_lon, lev)float320.0 0.0 0.0 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- pom_c1 in cloud water
- basename :
- pom_c1
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - pom_c3(time_lat_lon, lev)float320.0 0.0 0.0 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- pom_c3 in cloud water
- basename :
- pom_c3
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - pom_c4(time_lat_lon, lev)float320.0 0.0 0.0 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- pom_c4 in cloud water
- basename :
- pom_c4
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - so4_a1(time_lat_lon, lev)float329.182e-11 9.185e-11 ... 5.673e-21
- mdims :
- 9
- units :
- kg/kg
- long_name :
- so4_a1 concentration
- basename :
- so4_a1
array([[9.1824826e-11, 9.1849243e-11, 9.1898836e-11, ..., 2.7273122e-19, 2.7704460e-20, 1.7752790e-20], [9.3611244e-11, 9.3614880e-11, 9.3626086e-11, ..., 2.7434926e-19, 2.4224899e-20, 1.8572014e-20], [9.6905831e-11, 9.6899815e-11, 9.6891717e-11, ..., 3.0194631e-19, 2.6368267e-20, 2.1441097e-20], ..., [7.7908055e-11, 7.7899132e-11, 7.7831006e-11, ..., 1.7366586e-19, 1.6803725e-20, 5.3327635e-21], [8.7949911e-11, 8.7672168e-11, 8.6835351e-11, ..., 2.1465668e-19, 1.6267452e-20, 5.4017172e-21], [8.9691810e-11, 8.9237999e-11, 8.7905010e-11, ..., 2.2456700e-19, 1.4465773e-20, 5.6730256e-21]], dtype=float32) - so4_a2(time_lat_lon, lev)float322.249e-12 2.248e-12 ... 2.748e-14
- mdims :
- 9
- units :
- kg/kg
- long_name :
- so4_a2 concentration
- basename :
- so4_a2
array([[2.2489241e-12, 2.2477296e-12, 2.2451494e-12, ..., 4.2287100e-14, 3.5077267e-14, 3.3943937e-14], [2.2999660e-12, 2.2988605e-12, 2.2964172e-12, ..., 4.2267703e-14, 3.6659986e-14, 3.4156695e-14], [2.3849449e-12, 2.3840621e-12, 2.3820047e-12, ..., 4.3838234e-14, 3.6961889e-14, 3.2760443e-14], ..., [4.0156220e-12, 3.9985450e-12, 3.9467444e-12, ..., 3.4760199e-14, 3.3624694e-14, 3.0032976e-14], [3.9687988e-12, 3.9484874e-12, 3.8897284e-12, ..., 3.5191132e-14, 3.2568468e-14, 2.8929784e-14], [3.6872012e-12, 3.6549284e-12, 3.5623123e-12, ..., 3.5525317e-14, 3.1647041e-14, 2.7477612e-14]], dtype=float32) - so4_a3(time_lat_lon, lev)float324.968e-12 5.036e-12 ... 8.885e-32
- mdims :
- 9
- units :
- kg/kg
- long_name :
- so4_a3 concentration
- basename :
- so4_a3
array([[4.9676010e-12, 5.0364010e-12, 5.3381214e-12, ..., 2.2347453e-28, 1.1949160e-31, 8.9193590e-32], [4.9056783e-12, 4.9602930e-12, 5.2209153e-12, ..., 2.0665849e-28, 1.1098987e-31, 8.4957097e-32], [1.0395829e-11, 1.1031414e-11, 1.1017325e-11, ..., 1.9835581e-28, 1.3566956e-31, 9.5571205e-32], ..., [2.8116650e-12, 2.8036375e-12, 2.7787859e-12, ..., 7.0507681e-29, 6.5597445e-32, 8.7182214e-32], [2.3311596e-12, 2.3212116e-12, 2.2922814e-12, ..., 8.4416976e-29, 6.5620808e-32, 8.8008698e-32], [1.8497714e-12, 1.8293791e-12, 1.7769534e-12, ..., 9.5500492e-29, 7.6401614e-32, 8.8850428e-32]], dtype=float32) - so4_c1(time_lat_lon, lev)float323.976e-20 3.975e-20 ... 3.974e-20
- mdims :
- 9
- units :
- kg/kg
- long_name :
- so4_c1 in cloud water
- basename :
- so4_c1
array([[3.9757247e-20, 3.9745738e-20, 3.9741805e-20, ..., 3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9757939e-20, 3.9745563e-20, 3.9741834e-20, ..., 3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9764495e-20, 3.9747602e-20, ..., 3.9738778e-20, 3.9738778e-20, 3.9738778e-20], ..., [3.9738778e-20, 3.9738778e-20, 3.9738778e-20, ..., 3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20, ..., 3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20, ..., 3.9738778e-20, 3.9738778e-20, 3.9738778e-20]], dtype=float32) - so4_c2(time_lat_lon, lev)float323.972e-20 3.973e-20 ... 3.974e-20
- mdims :
- 9
- units :
- kg/kg
- long_name :
- so4_c2 in cloud water
- basename :
- so4_c2
array([[3.9720305e-20, 3.9731814e-20, 3.9735750e-20, ..., 3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9719617e-20, 3.9731989e-20, 3.9735718e-20, ..., 3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9713058e-20, 3.9729953e-20, ..., 3.9738778e-20, 3.9738778e-20, 3.9738778e-20], ..., [3.9738778e-20, 3.9738778e-20, 3.9738778e-20, ..., 3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20, ..., 3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20, ..., 3.9738778e-20, 3.9738778e-20, 3.9738778e-20]], dtype=float32) - so4_c3(time_lat_lon, lev)float323.974e-20 3.974e-20 ... 3.974e-20
- mdims :
- 9
- units :
- kg/kg
- long_name :
- so4_c3 in cloud water
- basename :
- so4_c3
array([[3.9738778e-20, 3.9738778e-20, 3.9738778e-20, ..., 3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20, ..., 3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20, ..., 3.9738778e-20, 3.9738778e-20, 3.9738778e-20], ..., [3.9738778e-20, 3.9738778e-20, 3.9738778e-20, ..., 3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20, ..., 3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20, ..., 3.9738778e-20, 3.9738778e-20, 3.9738778e-20]], dtype=float32) - soa_a1(time_lat_lon, lev)float328.4e-11 8.406e-11 ... 3.894e-22
- mdims :
- 9
- units :
- kg/kg
- long_name :
- soa_a1 concentration
- basename :
- soa_a1
array([[8.3998794e-11, 8.4059676e-11, 8.4181905e-11, ..., 1.8600004e-20, 1.5750846e-21, 1.0118312e-21], [8.5242306e-11, 8.5261853e-11, 8.5308705e-11, ..., 1.8692753e-20, 1.3136499e-21, 1.0812415e-21], [8.9315756e-11, 8.9306625e-11, 8.9300338e-11, ..., 2.0587235e-20, 1.4854881e-21, 1.2573992e-21], ..., [1.3465097e-10, 1.3456922e-10, 1.3426402e-10, ..., 1.5303231e-20, 1.2308843e-21, 3.2773596e-22], [1.2157757e-10, 1.2135370e-10, 1.2065240e-10, ..., 1.8847897e-20, 1.2235926e-21, 3.6082437e-22], [1.0138498e-10, 1.0096197e-10, 9.9686315e-11, ..., 1.9581011e-20, 1.0801951e-21, 3.8942738e-22]], dtype=float32) - soa_a2(time_lat_lon, lev)float324.754e-13 4.782e-13 ... 1.923e-16
- mdims :
- 9
- units :
- kg/kg
- long_name :
- soa_a2 concentration
- basename :
- soa_a2
array([[4.7541403e-13, 4.7819810e-13, 4.8368449e-13, ..., 4.1459327e-16, 2.7734572e-16, 2.6725300e-16], [4.8255139e-13, 4.8462184e-13, 4.8878940e-13, ..., 3.9816960e-16, 2.9408383e-16, 2.7131405e-16], [5.2034820e-13, 5.2180021e-13, 5.2479993e-13, ..., 4.0613790e-16, 2.9579232e-16, 2.5562356e-16], ..., [2.0410774e-12, 2.0431471e-12, 2.0477188e-12, ..., 2.6527158e-16, 2.3998463e-16, 1.9729151e-16], [1.7207035e-12, 1.7210130e-12, 1.7201555e-12, ..., 2.7624754e-16, 2.3123412e-16, 1.9468123e-16], [1.3332078e-12, 1.3293448e-12, 1.3160651e-12, ..., 2.8008584e-16, 2.2444970e-16, 1.9227365e-16]], dtype=float32) - soa_a3(time_lat_lon, lev)float325.266e-12 5.328e-12 ... 1.005e-36
- mdims :
- 9
- units :
- kg/kg
- long_name :
- soa_a3 concentration
- basename :
- soa_a3
array([[5.26648690e-12, 5.32790955e-12, 5.59568798e-12, ..., 1.00000004e-36, 1.00050558e-36, 1.00000004e-36], [5.17024141e-12, 5.21737211e-12, 5.44431817e-12, ..., 1.00003950e-36, 1.00019653e-36, 1.00000004e-36], [1.00755550e-11, 1.06348845e-11, 1.06198219e-11, ..., 1.00000748e-36, 1.00021205e-36, 1.00369516e-36], ..., [6.54148610e-12, 6.52777138e-12, 6.48416997e-12, ..., 1.00000004e-36, 1.00000004e-36, 1.00878010e-36], [5.32845426e-12, 5.31239679e-12, 5.26438051e-12, ..., 1.00000004e-36, 1.00000004e-36, 1.00836855e-36], [4.11106088e-12, 4.07655984e-12, 3.98181878e-12, ..., 1.00000004e-36, 1.00000004e-36, 1.00483189e-36]], dtype=float32) - soa_c1(time_lat_lon, lev)float320.0 0.0 0.0 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- soa_c1 in cloud water
- basename :
- soa_c1
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - soa_c2(time_lat_lon, lev)float320.0 0.0 0.0 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- soa_c2 in cloud water
- basename :
- soa_c2
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - soa_c3(time_lat_lon, lev)float320.0 0.0 0.0 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- soa_c3 in cloud water
- basename :
- soa_c3
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], dtype=float32) - wat_a1(time_lat_lon, lev)float323.214e-10 3.573e-10 ... 0.0 0.0
- mdims :
- 9
- units :
- m
- long_name :
- aerosol water, interstitial, mode 01
- basename :
- wat_a1
array([[3.2135011e-10, 3.5732087e-10, 4.4127707e-10, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [3.0491307e-10, 3.3728759e-10, 4.1163994e-10, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [2.9162270e-10, 3.2130118e-10, 3.8799089e-10, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [5.1843702e-10, 5.8133437e-10, 7.0895517e-10, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [5.9329708e-10, 6.8736827e-10, 9.0682289e-10, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [6.1191863e-10, 7.2556222e-10, 1.0189177e-09, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], dtype=float32) - wat_a2(time_lat_lon, lev)float323.821e-12 4.221e-12 ... 0.0 0.0
- mdims :
- 9
- units :
- m
- long_name :
- aerosol water, interstitial, mode 02
- basename :
- wat_a2
array([[3.8205884e-12, 4.2214674e-12, 5.1398517e-12, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [3.6551075e-12, 4.0201818e-12, 4.8443840e-12, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [3.5293825e-12, 3.8685964e-12, 4.6189879e-12, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [9.8964795e-12, 1.0975607e-11, 1.3075133e-11, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [1.0566703e-11, 1.2072635e-11, 1.5433176e-11, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [1.0220330e-11, 1.1887657e-11, 1.5908200e-11, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], dtype=float32) - wat_a3(time_lat_lon, lev)float322.606e-09 2.898e-09 ... 0.0 0.0
- mdims :
- 9
- units :
- m
- long_name :
- aerosol water, interstitial, mode 03
- basename :
- wat_a3
array([[2.6057103e-09, 2.8976659e-09, 3.5829792e-09, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [2.2657052e-09, 2.5066720e-09, 3.0632161e-09, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [2.0048059e-09, 2.2079187e-09, 2.6668405e-09, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [3.6754093e-09, 4.1356443e-09, 5.0754103e-09, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [3.7277013e-09, 4.3424060e-09, 5.7985816e-09, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [3.5761822e-09, 4.2635810e-09, 6.0637992e-09, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], dtype=float32) - wat_a4(time_lat_lon, lev)float321.733e-17 1.919e-17 ... 0.0 0.0
- mdims :
- 9
- units :
- m
- long_name :
- aerosol water, interstitial, mode 04
- basename :
- wat_a4
array([[1.7331460e-17, 1.9185771e-17, 2.3420984e-17, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [1.6853577e-17, 1.8572508e-17, 2.2443772e-17, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [1.6067368e-17, 1.7645594e-17, 2.1130507e-17, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [3.9561802e-17, 4.4129609e-17, 5.3452818e-17, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [4.8077947e-17, 5.5221605e-17, 7.1585673e-17, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [5.5252065e-17, 6.4844104e-17, 8.8830741e-17, ..., 0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], dtype=float32) - p_3d(time_lat_lon, lev)float64dask.array<chunksize=(72, 72), meta=np.ndarray>
- units :
- hPa
Array Chunk Bytes 40.50 kiB 40.50 kiB Shape (72, 72) (72, 72) Count 15 Tasks 1 Chunks Type float64 numpy.ndarray - zeros_cf(time_lat_lon, lev)float640.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- long_name :
- An array of zeros as only strat output is used for this model
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]]) - rho_a(time_lat_lon, lev)float64dask.array<chunksize=(72, 72), meta=np.ndarray>
- units :
- kg / m ** 3
Array Chunk Bytes 40.50 kiB 40.50 kiB Shape (72, 72) (72, 72) Count 18 Tasks 1 Chunks Type float64 numpy.ndarray
- Conventions :
- CF-1.0
- source :
- CAM
- case :
- EAMv1_ARM_Freerun_1
- title :
- UNSET
- logname :
- yshi
- host :
- cori07
- Version :
- $Name$
- revision_Id :
- $Id$
- initial_file :
- /global/cfs/cdirs/e3sm/inputdata/atm/cam/inic/homme/cami_mam3_Linoz_ne30np4_L72_c160214.nc
- topography_file :
- /global/cfs/cdirs/e3sm/inputdata/atm/cam/topo/USGS-gtopo30_ne30np4_16xdel2-PFC-consistentSGH.nc
- time_period_freq :
- hour_1
- _file_dates :
- ['20121031']
- _file_times :
- ['010000']
- _datastream :
- act_datastream
- _arm_standards_flag :
- 0
Running the Subcolumn Generator and Instrument Simulator#
Now, we use the emc2.simulator.main.make_simulated_data method to have EMC² perform a set of (optional) tasks:
Generate user-specified number of subcolumns per model grid cell.
Run the instrument simualtor.
Classify hydrometeor phase using the simulated observables.
We first call the emc2.simulator.main.make_simulated_data method to calculate the HSRL observables and then call it again to calcualte the KAZR observables (without invoking the subcolumn generator methods again).
Note that because the model output contains the spatial dimension stacked onto the time dimension, we wish to unstack these dimensions once all simualtor operations are done by setting unstack_dims=True.
because the simulator is being run here using a 10-yr old machine, the processing timing printed on the screen is relatively longer than one should typically expect.
#Specify number of subcolumns and run simulator (first, for the lidar, then the radar)
N_sub = 50
my_e3sm = emc2.simulator.main.make_simulated_data(my_e3sm, HSRL, N_sub, do_classify=True, convert_zeros_to_nan=True)
print("lidar processing done!")
my_model = emc2.simulator.main.make_simulated_data(my_e3sm, KAZR, N_sub, do_classify=True, convert_zeros_to_nan=True,
unstack_dims=True, skip_subcol_gen=True, finalize_fields=True)
print("radar processing done!")
## Creating subcolumns...
No convective processing for E3SM
Now performing parallel stratiform hydrometeor allocation in subcolumns
Fully overcast cl & ci in 649 voxels
Done! total processing time = 7.33s
Now performing parallel strat precipitation allocation in subcolumns
Fully overcast pl & pi in 420 voxels
Done! total processing time = 8.80s
Generating lidar moments...
Generating stratiform lidar variables using radiation logic
2-D interpolation of bulk liq lidar backscattering using mu-lambda values
2-D interpolation of bulk liq lidar extinction using mu-lambda values
Done! total processing time = 0.55s
'pl' not in hyd_types = ['cl', 'ci', 'pi']; excluding from COSP calculations
lidar processing done!
## Creating subcolumns...
Skipping subcolumn generator (make sure subcolumns were already generated).
Generating radar moments...
Generating stratiform radar variables using radiation logic
2-D interpolation of bulk liq radar backscattering using mu-lambda values
2-D interpolation of bulk liq radar extinction using mu-lambda values
Done! total processing time = 0.42s
Unstacking the time dimension (time, lat, and lon dimensions)
radar processing done!
We can examine the simulator output dataset saved under an xr.Dataset object. We can see that for most lidar and radar observables (the equivalent reflectivity factor Ze, the backscatter cross-section beta_p, etc.) we have fields per hydrometeor class in addition to the field representing the total.#
my_e3sm.ds
<xarray.Dataset>
Dimensions: (ncol: 3, time: 24, lev: 72,
subcolumn: 50, ilev: 73, cosp_prs: 7,
nbnd: 2, cosp_tau: 7, cosp_scol: 10,
cosp_ht: 40, cosp_sr: 15, cosp_sza: 5,
cosp_htmisr: 16, cosp_tau_modis: 6)
Coordinates: (12/13)
* ncol (ncol) int64 0 1 2
* time (time) datetime64[ns] 2012-10-31T01:00:...
* lev (lev) float64 998.5 993.8 ... 0.1238
* subcolumn (subcolumn) int64 0 1 2 3 ... 46 47 48 49
* ilev (ilev) float64 0.1 0.1477 ... 997.0 1e+03
* cosp_prs (cosp_prs) float64 900.0 740.0 ... 90.0
... ...
* cosp_scol (cosp_scol) int32 1 2 3 4 5 6 7 8 9 10
* cosp_ht (cosp_ht) float64 240.0 ... 1.896e+04
* cosp_sr (cosp_sr) float64 0.605 2.1 ... 1.004e+03
* cosp_sza (cosp_sza) float64 0.0 15.0 30.0 45.0 60.0
* cosp_htmisr (cosp_htmisr) float64 -99.0 0.25 ... 58.0
* cosp_tau_modis (cosp_tau_modis) float64 0.8 ... 5.003e+04
Dimensions without coordinates: nbnd
Data variables: (12/268)
lat (time, ncol) float64 72.89 71.18 ... 70.44
lon (time, ncol) float64 202.9 204.9 ... 203.8
cosp_prs_bnds (cosp_prs, nbnd) float64 dask.array<chunksize=(7, 2), meta=np.ndarray>
cosp_tau_bnds (nbnd, cosp_tau) float64 dask.array<chunksize=(2, 7), meta=np.ndarray>
cosp_ht_bnds (nbnd, cosp_ht) float64 dask.array<chunksize=(2, 40), meta=np.ndarray>
cosp_sr_bnds (nbnd, cosp_sr) float64 dask.array<chunksize=(2, 15), meta=np.ndarray>
... ...
sub_col_Ze_att_tot_strat (subcolumn, time, lev, ncol) float64 na...
Ze_min (time, lev, ncol) float32 -87.04 ... -1...
sub_col_Ze_tot (subcolumn, time, lev, ncol) float64 na...
sub_col_Ze_att_tot (subcolumn, time, lev, ncol) float64 na...
detect_mask (subcolumn, time, lev, ncol) bool False...
phase_mask_KAZR_sounding_all_hyd (subcolumn, time, lev, ncol) float64 na...
Attributes: (12/15)
Conventions: CF-1.0
source: CAM
case: EAMv1_ARM_Freerun_1
title: UNSET
logname: yshi
host: cori07
... ...
topography_file: /global/cfs/cdirs/e3sm/inputdata/atm/cam/topo/USGS-...
time_period_freq: hour_1
_file_dates: ['20121031']
_file_times: ['010000']
_datastream: act_datastream
_arm_standards_flag: 0- ncol: 3
- time: 24
- lev: 72
- subcolumn: 50
- ilev: 73
- cosp_prs: 7
- nbnd: 2
- cosp_tau: 7
- cosp_scol: 10
- cosp_ht: 40
- cosp_sr: 15
- cosp_sza: 5
- cosp_htmisr: 16
- cosp_tau_modis: 6
- ncol(ncol)int640 1 2
array([0, 1, 2])
- time(time)datetime64[ns]2012-10-31T01:00:00 ... 2012-11-01
array(['2012-10-31T01:00:00.000000000', '2012-10-31T02:00:00.000000000', '2012-10-31T03:00:00.000000000', '2012-10-31T04:00:00.000000000', '2012-10-31T05:00:00.000000000', '2012-10-31T06:00:00.000000000', '2012-10-31T07:00:00.000000000', '2012-10-31T08:00:00.000000000', '2012-10-31T09:00:00.000000000', '2012-10-31T10:00:00.000000000', '2012-10-31T11:00:00.000000000', '2012-10-31T12:00:00.000000000', '2012-10-31T13:00:00.000000000', '2012-10-31T14:00:00.000000000', '2012-10-31T15:00:00.000000000', '2012-10-31T16:00:00.000000000', '2012-10-31T17:00:00.000000000', '2012-10-31T18:00:00.000000000', '2012-10-31T19:00:00.000000000', '2012-10-31T20:00:00.000000000', '2012-10-31T21:00:00.000000000', '2012-10-31T22:00:00.000000000', '2012-10-31T23:00:00.000000000', '2012-11-01T00:00:00.000000000'], dtype='datetime64[ns]') - lev(lev)float64998.5 993.8 986.2 ... 0.1828 0.1238
array([9.984964e+02, 9.937570e+02, 9.862053e+02, 9.773141e+02, 9.679111e+02, 9.580128e+02, 9.476362e+02, 9.367990e+02, 9.255197e+02, 9.138175e+02, 9.017118e+02, 8.892227e+02, 8.763706e+02, 8.631764e+02, 8.496612e+02, 8.350952e+02, 8.178688e+02, 7.969527e+02, 7.723628e+02, 7.444748e+02, 7.137046e+02, 6.804980e+02, 6.453200e+02, 6.086438e+02, 5.706249e+02, 5.316395e+02, 4.924686e+02, 4.543958e+02, 4.188035e+02, 3.859990e+02, 3.557641e+02, 3.278975e+02, 3.022136e+02, 2.785416e+02, 2.567237e+02, 2.366148e+02, 2.180810e+02, 2.009989e+02, 1.852549e+02, 1.707441e+02, 1.573699e+02, 1.450433e+02, 1.336822e+02, 1.232110e+02, 1.135600e+02, 1.046650e+02, 9.646667e+01, 8.891054e+01, 8.194628e+01, 7.535534e+01, 6.847639e+01, 6.101518e+01, 5.330956e+01, 4.567120e+01, 3.836628e+01, 3.159325e+01, 2.544470e+01, 1.999634e+01, 1.533394e+01, 1.147379e+01, 8.377404e+00, 5.968449e+00, 4.129833e+00, 2.797027e+00, 1.894352e+00, 1.282995e+00, 8.689386e-01, 5.885091e-01, 3.985817e-01, 2.699489e-01, 1.828292e-01, 1.238254e-01]) - subcolumn(subcolumn)int640 1 2 3 4 5 6 ... 44 45 46 47 48 49
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49]) - ilev(ilev)float640.1 0.1477 0.218 ... 997.0 1e+03
array([1.000000e-01, 1.476508e-01, 2.180076e-01, 3.218901e-01, 4.752733e-01, 7.017450e-01, 1.036132e+00, 1.529858e+00, 2.258847e+00, 3.335207e+00, 4.924460e+00, 7.012439e+00, 9.742370e+00, 1.320520e+01, 1.746267e+01, 2.253000e+01, 2.835939e+01, 3.482711e+01, 4.190545e+01, 4.943694e+01, 5.718218e+01, 6.484818e+01, 7.210460e+01, 7.860608e+01, 8.528648e+01, 9.253461e+01, 1.003987e+02, 1.089312e+02, 1.181888e+02, 1.282332e+02, 1.391312e+02, 1.509554e+02, 1.637844e+02, 1.777038e+02, 1.928061e+02, 2.091918e+02, 2.269702e+02, 2.462594e+02, 2.671880e+02, 2.898952e+02, 3.145321e+02, 3.412629e+02, 3.702654e+02, 4.017327e+02, 4.358743e+02, 4.729174e+02, 5.120198e+02, 5.512593e+02, 5.899905e+02, 6.272970e+02, 6.633429e+02, 6.976532e+02, 7.297561e+02, 7.591936e+02, 7.855321e+02, 8.083734e+02, 8.273643e+02, 8.428261e+02, 8.564964e+02, 8.698564e+02, 8.828849e+02, 8.955606e+02, 9.078631e+02, 9.197720e+02, 9.312675e+02, 9.423305e+02, 9.529419e+02, 9.630837e+02, 9.727385e+02, 9.818896e+02, 9.905210e+02, 9.969929e+02, 1.000000e+03]) - cosp_prs(cosp_prs)float64900.0 740.0 620.0 ... 245.0 90.0
- long_name :
- COSP Mean ISCCP pressure
- units :
- hPa
- bounds :
- cosp_prs_bnds
array([900., 740., 620., 500., 375., 245., 90.])
- cosp_tau(cosp_tau)float640.15 0.8 2.45 6.5 16.2 41.5 219.5
- long_name :
- COSP Mean ISCCP optical depth
- units :
- 1
- bounds :
- cosp_tau_bnds
array([1.500e-01, 8.000e-01, 2.450e+00, 6.500e+00, 1.620e+01, 4.150e+01, 2.195e+02]) - cosp_scol(cosp_scol)int321 2 3 4 5 6 7 8 9 10
- long_name :
- COSP subcolumn
array([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10], dtype=int32)
- cosp_ht(cosp_ht)float64240.0 720.0 ... 1.848e+04 1.896e+04
- long_name :
- COSP Mean Height for lidar and radar simulator outputs
- units :
- m
- bounds :
- cosp_ht_bnds
array([ 240., 720., 1200., 1680., 2160., 2640., 3120., 3600., 4080., 4560., 5040., 5520., 6000., 6480., 6960., 7440., 7920., 8400., 8880., 9360., 9840., 10320., 10800., 11280., 11760., 12240., 12720., 13200., 13680., 14160., 14640., 15120., 15600., 16080., 16560., 17040., 17520., 18000., 18480., 18960.]) - cosp_sr(cosp_sr)float640.605 2.1 4.0 ... 539.5 1.004e+03
- long_name :
- COSP Mean Scattering Ratio for lidar simulator CFAD output
- units :
- 1
- bounds :
- cosp_sr_bnds
array([6.050e-01, 2.100e+00, 4.000e+00, 6.000e+00, 8.500e+00, 1.250e+01, 1.750e+01, 2.250e+01, 2.750e+01, 3.500e+01, 4.500e+01, 5.500e+01, 7.000e+01, 5.395e+02, 1.004e+03]) - cosp_sza(cosp_sza)float640.0 15.0 30.0 45.0 60.0
- long_name :
- COSP Parasol SZA
- units :
- degrees
array([ 0., 15., 30., 45., 60.])
- cosp_htmisr(cosp_htmisr)float64-99.0 0.25 0.75 ... 14.0 16.0 58.0
- long_name :
- COSP MISR height
- units :
- km
- bounds :
- cosp_htmisr_bnds
array([-99. , 0.25, 0.75, 1.25, 1.75, 2.25, 2.75, 3.5 , 4.5 , 6. , 8. , 10. , 12. , 14. , 16. , 58. ]) - cosp_tau_modis(cosp_tau_modis)float640.8 2.45 6.5 16.2 41.5 5.003e+04
- long_name :
- COSP Mean MODIS optical depth
- units :
- 1
- bounds :
- cosp_tau_modis_bnds
array([8.000e-01, 2.450e+00, 6.500e+00, 1.620e+01, 4.150e+01, 5.003e+04])
- lat(time, ncol)float6472.89 71.18 70.44 ... 71.18 70.44
- long_name :
- latitude
- units :
- degrees_north
array([[72.89282783, 71.18477818, 70.44340981], [72.89282783, 71.18477818, 70.44340981], [72.89282783, 71.18477818, 70.44340981], [72.89282783, 71.18477818, 70.44340981], [72.89282783, 71.18477818, 70.44340981], [72.89282783, 71.18477818, 70.44340981], [72.89282783, 71.18477818, 70.44340981], [72.89282783, 71.18477818, 70.44340981], [72.89282783, 71.18477818, 70.44340981], [72.89282783, 71.18477818, 70.44340981], [72.89282783, 71.18477818, 70.44340981], [72.89282783, 71.18477818, 70.44340981], [72.89282783, 71.18477818, 70.44340981], [72.89282783, 71.18477818, 70.44340981], [72.89282783, 71.18477818, 70.44340981], [72.89282783, 71.18477818, 70.44340981], [72.89282783, 71.18477818, 70.44340981], [72.89282783, 71.18477818, 70.44340981], [72.89282783, 71.18477818, 70.44340981], [72.89282783, 71.18477818, 70.44340981], [72.89282783, 71.18477818, 70.44340981], [72.89282783, 71.18477818, 70.44340981], [72.89282783, 71.18477818, 70.44340981], [72.89282783, 71.18477818, 70.44340981]]) - lon(time, ncol)float64202.9 204.9 203.8 ... 204.9 203.8
- long_name :
- longitude
- units :
- degrees_east
array([[202.89916211, 204.92322624, 203.84068991], [202.89916211, 204.92322624, 203.84068991], [202.89916211, 204.92322624, 203.84068991], [202.89916211, 204.92322624, 203.84068991], [202.89916211, 204.92322624, 203.84068991], [202.89916211, 204.92322624, 203.84068991], [202.89916211, 204.92322624, 203.84068991], [202.89916211, 204.92322624, 203.84068991], [202.89916211, 204.92322624, 203.84068991], [202.89916211, 204.92322624, 203.84068991], [202.89916211, 204.92322624, 203.84068991], [202.89916211, 204.92322624, 203.84068991], [202.89916211, 204.92322624, 203.84068991], [202.89916211, 204.92322624, 203.84068991], [202.89916211, 204.92322624, 203.84068991], [202.89916211, 204.92322624, 203.84068991], [202.89916211, 204.92322624, 203.84068991], [202.89916211, 204.92322624, 203.84068991], [202.89916211, 204.92322624, 203.84068991], [202.89916211, 204.92322624, 203.84068991], [202.89916211, 204.92322624, 203.84068991], [202.89916211, 204.92322624, 203.84068991], [202.89916211, 204.92322624, 203.84068991], [202.89916211, 204.92322624, 203.84068991]]) - cosp_prs_bnds(cosp_prs, nbnd)float64dask.array<chunksize=(7, 2), meta=np.ndarray>
Array Chunk Bytes 112 B 112 B Shape (7, 2) (7, 2) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - cosp_tau_bnds(nbnd, cosp_tau)float64dask.array<chunksize=(2, 7), meta=np.ndarray>
Array Chunk Bytes 112 B 112 B Shape (2, 7) (2, 7) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - cosp_ht_bnds(nbnd, cosp_ht)float64dask.array<chunksize=(2, 40), meta=np.ndarray>
Array Chunk Bytes 640 B 640 B Shape (2, 40) (2, 40) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - cosp_sr_bnds(nbnd, cosp_sr)float64dask.array<chunksize=(2, 15), meta=np.ndarray>
Array Chunk Bytes 240 B 240 B Shape (2, 15) (2, 15) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - cosp_htmisr_bnds(nbnd, cosp_htmisr)float64dask.array<chunksize=(2, 16), meta=np.ndarray>
Array Chunk Bytes 256 B 256 B Shape (2, 16) (2, 16) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - cosp_tau_modis_bnds(nbnd, cosp_tau_modis)float64dask.array<chunksize=(2, 6), meta=np.ndarray>
Array Chunk Bytes 96 B 96 B Shape (2, 6) (2, 6) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - hyam(lev)float640.0 0.0 ... 0.0001828 0.0001238
- long_name :
- hybrid A coefficient at layer midpoints
array([0.00000000e+00, 0.00000000e+00, 3.64808452e-04, 1.76556810e-03, 3.89449073e-03, 6.13557040e-03, 8.48494102e-03, 1.09385772e-02, 1.34923049e-02, 1.61418126e-02, 1.88826608e-02, 2.17103133e-02, 2.46201493e-02, 2.76074433e-02, 3.06674030e-02, 3.39653047e-02, 3.78655213e-02, 4.26011286e-02, 4.81685284e-02, 5.44826500e-02, 6.14493199e-02, 6.89676092e-02, 7.69322627e-02, 8.52361112e-02, 9.38439551e-02, 1.02670630e-01, 1.11539324e-01, 1.20159364e-01, 1.28217812e-01, 1.35645058e-01, 1.42490535e-01, 1.48799804e-01, 1.54614873e-01, 1.59974463e-01, 1.64914246e-01, 1.69467100e-01, 1.73663333e-01, 1.77530875e-01, 1.78544788e-01, 1.70744080e-01, 1.57369878e-01, 1.45043266e-01, 1.33682180e-01, 1.23210991e-01, 1.13560001e-01, 1.04664967e-01, 9.64666734e-02, 8.89105431e-02, 8.19462751e-02, 7.53553359e-02, 6.84763902e-02, 6.10151817e-02, 5.33095614e-02, 4.56711979e-02, 3.83662831e-02, 3.15932513e-02, 2.54446965e-02, 1.99963380e-02, 1.53339382e-02, 1.14737872e-02, 8.37740438e-03, 5.96844937e-03, 4.12983316e-03, 2.79702694e-03, 1.89435248e-03, 1.28299491e-03, 8.68938570e-04, 5.88509147e-04, 3.98581704e-04, 2.69948862e-04, 1.82829236e-04, 1.23825413e-04]) - hybm(lev)float640.9985 0.9938 0.9858 ... 0.0 0.0
- long_name :
- hybrid B coefficient at layer midpoints
array([0.99849644, 0.99375696, 0.98584052, 0.9755485 , 0.96401659, 0.9518772 , 0.93915122, 0.92586042, 0.91202744, 0.89767569, 0.88282917, 0.86751242, 0.8517505 , 0.83556898, 0.81899385, 0.80112987, 0.7800033 , 0.75435162, 0.72419431, 0.68999218, 0.65225532, 0.61153044, 0.56838771, 0.52340766, 0.47678095, 0.4289689 , 0.38092925, 0.33423645, 0.29058565, 0.25035397, 0.21327359, 0.1790977 , 0.14759877, 0.1185671 , 0.09180944, 0.06714769, 0.04441766, 0.02346807, 0.00671011, 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ]) - P0()float64...
- long_name :
- reference pressure
- units :
- Pa
array(100000.)
- hyai(ilev)float64dask.array<chunksize=(73,), meta=np.ndarray>
- long_name :
- hybrid A coefficient at layer interfaces
Array Chunk Bytes 584 B 584 B Shape (73,) (73,) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - hybi(ilev)float64dask.array<chunksize=(73,), meta=np.ndarray>
- long_name :
- hybrid B coefficient at layer interfaces
Array Chunk Bytes 584 B 584 B Shape (73,) (73,) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - date(time, ncol)int3220121031 20121031 ... 20121101
- long_name :
- current date (YYYYMMDD)
array([[20121031, 20121031, 20121031], [20121031, 20121031, 20121031], [20121031, 20121031, 20121031], [20121031, 20121031, 20121031], [20121031, 20121031, 20121031], [20121031, 20121031, 20121031], [20121031, 20121031, 20121031], [20121031, 20121031, 20121031], [20121031, 20121031, 20121031], [20121031, 20121031, 20121031], [20121031, 20121031, 20121031], [20121031, 20121031, 20121031], [20121031, 20121031, 20121031], [20121031, 20121031, 20121031], [20121031, 20121031, 20121031], [20121031, 20121031, 20121031], [20121031, 20121031, 20121031], [20121031, 20121031, 20121031], [20121031, 20121031, 20121031], [20121031, 20121031, 20121031], [20121031, 20121031, 20121031], [20121031, 20121031, 20121031], [20121031, 20121031, 20121031], [20121101, 20121101, 20121101]], dtype=int32) - datesec(time, ncol)int323600 3600 3600 7200 ... 82800 0 0 0
- long_name :
- current seconds of current date
array([[ 3600, 3600, 3600], [ 7200, 7200, 7200], [10800, 10800, 10800], [14400, 14400, 14400], [18000, 18000, 18000], [21600, 21600, 21600], [25200, 25200, 25200], [28800, 28800, 28800], [32400, 32400, 32400], [36000, 36000, 36000], [39600, 39600, 39600], [43200, 43200, 43200], [46800, 46800, 46800], [50400, 50400, 50400], [54000, 54000, 54000], [57600, 57600, 57600], [61200, 61200, 61200], [64800, 64800, 64800], [68400, 68400, 68400], [72000, 72000, 72000], [75600, 75600, 75600], [79200, 79200, 79200], [82800, 82800, 82800], [ 0, 0, 0]], dtype=int32) - time_bnds(time, ncol, nbnd)object2012-10-31 00:00:00 ... 2012-11-...
- long_name :
- time interval endpoints
array([[[cftime.DatetimeGregorian(2012, 10, 31, 0, 0, 0, 0, has_year_zero=False), cftime.DatetimeGregorian(2012, 10, 31, 1, 0, 0, 0, has_year_zero=False)], [cftime.DatetimeGregorian(2012, 10, 31, 0, 0, 0, 0, has_year_zero=False), cftime.DatetimeGregorian(2012, 10, 31, 1, 0, 0, 0, has_year_zero=False)], [cftime.DatetimeGregorian(2012, 10, 31, 0, 0, 0, 0, has_year_zero=False), cftime.DatetimeGregorian(2012, 10, 31, 1, 0, 0, 0, has_year_zero=False)]], [[cftime.DatetimeGregorian(2012, 10, 31, 1, 0, 0, 0, has_year_zero=False), cftime.DatetimeGregorian(2012, 10, 31, 2, 0, 0, 0, has_year_zero=False)], [cftime.DatetimeGregorian(2012, 10, 31, 1, 0, 0, 0, has_year_zero=False), cftime.DatetimeGregorian(2012, 10, 31, 2, 0, 0, 0, has_year_zero=False)], [cftime.DatetimeGregorian(2012, 10, 31, 1, 0, 0, 0, has_year_zero=False), cftime.DatetimeGregorian(2012, 10, 31, 2, 0, 0, 0, has_year_zero=False)]], [[cftime.DatetimeGregorian(2012, 10, 31, 2, 0, 0, 0, has_year_zero=False), cftime.DatetimeGregorian(2012, 10, 31, 3, 0, 0, 0, has_year_zero=False)], [cftime.DatetimeGregorian(2012, 10, 31, 2, 0, 0, 0, has_year_zero=False), cftime.DatetimeGregorian(2012, 10, 31, 3, 0, 0, 0, has_year_zero=False)], [cftime.DatetimeGregorian(2012, 10, 31, 2, 0, 0, 0, has_year_zero=False), cftime.DatetimeGregorian(2012, 10, 31, 3, 0, 0, 0, has_year_zero=False)]], ... cftime.DatetimeGregorian(2012, 10, 31, 22, 0, 0, 0, has_year_zero=False)], [cftime.DatetimeGregorian(2012, 10, 31, 21, 0, 0, 0, has_year_zero=False), cftime.DatetimeGregorian(2012, 10, 31, 22, 0, 0, 0, has_year_zero=False)], [cftime.DatetimeGregorian(2012, 10, 31, 21, 0, 0, 0, has_year_zero=False), cftime.DatetimeGregorian(2012, 10, 31, 22, 0, 0, 0, has_year_zero=False)]], [[cftime.DatetimeGregorian(2012, 10, 31, 22, 0, 0, 0, has_year_zero=False), cftime.DatetimeGregorian(2012, 10, 31, 23, 0, 0, 0, has_year_zero=False)], [cftime.DatetimeGregorian(2012, 10, 31, 22, 0, 0, 0, has_year_zero=False), cftime.DatetimeGregorian(2012, 10, 31, 23, 0, 0, 0, has_year_zero=False)], [cftime.DatetimeGregorian(2012, 10, 31, 22, 0, 0, 0, has_year_zero=False), cftime.DatetimeGregorian(2012, 10, 31, 23, 0, 0, 0, has_year_zero=False)]], [[cftime.DatetimeGregorian(2012, 10, 31, 23, 0, 0, 0, has_year_zero=False), cftime.DatetimeGregorian(2012, 11, 1, 0, 0, 0, 0, has_year_zero=False)], [cftime.DatetimeGregorian(2012, 10, 31, 23, 0, 0, 0, has_year_zero=False), cftime.DatetimeGregorian(2012, 11, 1, 0, 0, 0, 0, has_year_zero=False)], [cftime.DatetimeGregorian(2012, 10, 31, 23, 0, 0, 0, has_year_zero=False), cftime.DatetimeGregorian(2012, 11, 1, 0, 0, 0, 0, has_year_zero=False)]]], dtype=object) - date_written(time, ncol)|S8b'01/12/22' ... b'01/12/22'
array([[b'01/12/22', b'01/12/22', b'01/12/22'], [b'01/12/22', b'01/12/22', b'01/12/22'], [b'01/12/22', b'01/12/22', b'01/12/22'], [b'01/12/22', b'01/12/22', b'01/12/22'], [b'01/12/22', b'01/12/22', b'01/12/22'], [b'01/12/22', b'01/12/22', b'01/12/22'], [b'01/12/22', b'01/12/22', b'01/12/22'], [b'01/12/22', b'01/12/22', b'01/12/22'], [b'01/12/22', b'01/12/22', b'01/12/22'], [b'01/12/22', b'01/12/22', b'01/12/22'], [b'01/12/22', b'01/12/22', b'01/12/22'], [b'01/12/22', b'01/12/22', b'01/12/22'], [b'01/12/22', b'01/12/22', b'01/12/22'], [b'01/12/22', b'01/12/22', b'01/12/22'], [b'01/12/22', b'01/12/22', b'01/12/22'], [b'01/12/22', b'01/12/22', b'01/12/22'], [b'01/12/22', b'01/12/22', b'01/12/22'], [b'01/12/22', b'01/12/22', b'01/12/22'], [b'01/12/22', b'01/12/22', b'01/12/22'], [b'01/12/22', b'01/12/22', b'01/12/22'], [b'01/12/22', b'01/12/22', b'01/12/22'], [b'01/12/22', b'01/12/22', b'01/12/22'], [b'01/12/22', b'01/12/22', b'01/12/22'], [b'01/12/22', b'01/12/22', b'01/12/22']], dtype='|S8') - time_written(time, ncol)|S8b'00:04:38' ... b'00:08:17'
array([[b'00:04:38', b'00:04:38', b'00:04:38'], [b'00:04:46', b'00:04:46', b'00:04:46'], [b'00:04:53', b'00:04:53', b'00:04:53'], [b'00:05:00', b'00:05:00', b'00:05:00'], [b'00:05:07', b'00:05:07', b'00:05:07'], [b'00:05:14', b'00:05:14', b'00:05:14'], [b'00:05:21', b'00:05:21', b'00:05:21'], [b'00:05:28', b'00:05:28', b'00:05:28'], [b'00:05:35', b'00:05:35', b'00:05:35'], [b'00:05:42', b'00:05:42', b'00:05:42'], [b'00:05:49', b'00:05:49', b'00:05:49'], [b'00:05:56', b'00:05:56', b'00:05:56'], [b'00:06:03', b'00:06:03', b'00:06:03'], [b'00:06:10', b'00:06:10', b'00:06:10'], [b'00:06:17', b'00:06:17', b'00:06:17'], [b'00:06:24', b'00:06:24', b'00:06:24'], [b'00:06:30', b'00:06:30', b'00:06:30'], [b'00:06:38', b'00:06:38', b'00:06:38'], [b'00:06:45', b'00:06:45', b'00:06:45'], [b'00:06:52', b'00:06:52', b'00:06:52'], [b'00:06:59', b'00:06:59', b'00:06:59'], [b'00:07:06', b'00:07:06', b'00:07:06'], [b'00:07:12', b'00:07:12', b'00:07:12'], [b'00:08:17', b'00:08:17', b'00:08:17']], dtype='|S8') - ndbase()int32...
- long_name :
- base day
array(0, dtype=int32)
- nsbase()int32...
- long_name :
- seconds of base day
array(0, dtype=int32)
- nbdate()int32...
- long_name :
- base date (YYYYMMDD)
array(20110101, dtype=int32)
- nbsec()int32...
- long_name :
- seconds of base date
array(0, dtype=int32)
- mdt()int32...
- long_name :
- timestep
- units :
- s
array(1800, dtype=int32)
- ndcur(time, ncol)int32669 669 669 669 ... 669 670 670 670
- long_name :
- current day (from base day)
array([[669, 669, 669], [669, 669, 669], [669, 669, 669], [669, 669, 669], [669, 669, 669], [669, 669, 669], [669, 669, 669], [669, 669, 669], [669, 669, 669], [669, 669, 669], [669, 669, 669], [669, 669, 669], [669, 669, 669], [669, 669, 669], [669, 669, 669], [669, 669, 669], [669, 669, 669], [669, 669, 669], [669, 669, 669], [669, 669, 669], [669, 669, 669], [669, 669, 669], [669, 669, 669], [670, 670, 670]], dtype=int32) - nscur(time, ncol)int323600 3600 3600 7200 ... 82800 0 0 0
- long_name :
- current seconds of current day
array([[ 3600, 3600, 3600], [ 7200, 7200, 7200], [10800, 10800, 10800], [14400, 14400, 14400], [18000, 18000, 18000], [21600, 21600, 21600], [25200, 25200, 25200], [28800, 28800, 28800], [32400, 32400, 32400], [36000, 36000, 36000], [39600, 39600, 39600], [43200, 43200, 43200], [46800, 46800, 46800], [50400, 50400, 50400], [54000, 54000, 54000], [57600, 57600, 57600], [61200, 61200, 61200], [64800, 64800, 64800], [68400, 68400, 68400], [72000, 72000, 72000], [75600, 75600, 75600], [79200, 79200, 79200], [82800, 82800, 82800], [ 0, 0, 0]], dtype=int32) - co2vmr(time, ncol)float640.0003953 0.0003953 ... 0.0003953
- long_name :
- co2 volume mixing ratio
array([[0.00039527, 0.00039527, 0.00039527], [0.00039527, 0.00039527, 0.00039527], [0.00039527, 0.00039527, 0.00039527], [0.00039527, 0.00039527, 0.00039527], [0.00039527, 0.00039527, 0.00039527], [0.00039527, 0.00039527, 0.00039527], [0.00039527, 0.00039527, 0.00039527], [0.00039527, 0.00039527, 0.00039527], [0.00039528, 0.00039528, 0.00039528], [0.00039528, 0.00039528, 0.00039528], [0.00039528, 0.00039528, 0.00039528], [0.00039528, 0.00039528, 0.00039528], [0.00039528, 0.00039528, 0.00039528], [0.00039528, 0.00039528, 0.00039528], [0.00039528, 0.00039528, 0.00039528], [0.00039528, 0.00039528, 0.00039528], [0.00039528, 0.00039528, 0.00039528], [0.00039528, 0.00039528, 0.00039528], [0.00039528, 0.00039528, 0.00039528], [0.00039528, 0.00039528, 0.00039528], [0.00039528, 0.00039528, 0.00039528], [0.00039528, 0.00039528, 0.00039528], [0.00039528, 0.00039528, 0.00039528], [0.00039528, 0.00039528, 0.00039528]]) - ch4vmr(time, ncol)float641.821e-06 1.821e-06 ... 1.821e-06
- long_name :
- ch4 volume mixing ratio
array([[1.82135837e-06, 1.82135837e-06, 1.82135837e-06], [1.82135921e-06, 1.82135921e-06, 1.82135921e-06], [1.82136004e-06, 1.82136004e-06, 1.82136004e-06], [1.82136088e-06, 1.82136088e-06, 1.82136088e-06], [1.82136172e-06, 1.82136172e-06, 1.82136172e-06], [1.82136255e-06, 1.82136255e-06, 1.82136255e-06], [1.82136339e-06, 1.82136339e-06, 1.82136339e-06], [1.82136422e-06, 1.82136422e-06, 1.82136422e-06], [1.82136506e-06, 1.82136506e-06, 1.82136506e-06], [1.82136589e-06, 1.82136589e-06, 1.82136589e-06], [1.82136673e-06, 1.82136673e-06, 1.82136673e-06], [1.82136757e-06, 1.82136757e-06, 1.82136757e-06], [1.82136840e-06, 1.82136840e-06, 1.82136840e-06], [1.82136924e-06, 1.82136924e-06, 1.82136924e-06], [1.82137007e-06, 1.82137007e-06, 1.82137007e-06], [1.82137091e-06, 1.82137091e-06, 1.82137091e-06], [1.82137174e-06, 1.82137174e-06, 1.82137174e-06], [1.82137258e-06, 1.82137258e-06, 1.82137258e-06], [1.82137341e-06, 1.82137341e-06, 1.82137341e-06], [1.82137425e-06, 1.82137425e-06, 1.82137425e-06], [1.82137509e-06, 1.82137509e-06, 1.82137509e-06], [1.82137592e-06, 1.82137592e-06, 1.82137592e-06], [1.82137676e-06, 1.82137676e-06, 1.82137676e-06], [1.82137759e-06, 1.82137759e-06, 1.82137759e-06]]) - n2ovmr(time, ncol)float643.258e-07 3.258e-07 ... 3.258e-07
- long_name :
- n2o volume mixing ratio
array([[3.25766360e-07, 3.25766360e-07, 3.25766360e-07], [3.25766464e-07, 3.25766464e-07, 3.25766464e-07], [3.25766568e-07, 3.25766568e-07, 3.25766568e-07], [3.25766673e-07, 3.25766673e-07, 3.25766673e-07], [3.25766777e-07, 3.25766777e-07, 3.25766777e-07], [3.25766881e-07, 3.25766881e-07, 3.25766881e-07], [3.25766986e-07, 3.25766986e-07, 3.25766986e-07], [3.25767090e-07, 3.25767090e-07, 3.25767090e-07], [3.25767194e-07, 3.25767194e-07, 3.25767194e-07], [3.25767299e-07, 3.25767299e-07, 3.25767299e-07], [3.25767403e-07, 3.25767403e-07, 3.25767403e-07], [3.25767507e-07, 3.25767507e-07, 3.25767507e-07], [3.25767612e-07, 3.25767612e-07, 3.25767612e-07], [3.25767716e-07, 3.25767716e-07, 3.25767716e-07], [3.25767820e-07, 3.25767820e-07, 3.25767820e-07], [3.25767925e-07, 3.25767925e-07, 3.25767925e-07], [3.25768029e-07, 3.25768029e-07, 3.25768029e-07], [3.25768134e-07, 3.25768134e-07, 3.25768134e-07], [3.25768238e-07, 3.25768238e-07, 3.25768238e-07], [3.25768342e-07, 3.25768342e-07, 3.25768342e-07], [3.25768447e-07, 3.25768447e-07, 3.25768447e-07], [3.25768551e-07, 3.25768551e-07, 3.25768551e-07], [3.25768655e-07, 3.25768655e-07, 3.25768655e-07], [3.25768760e-07, 3.25768760e-07, 3.25768760e-07]]) - f11vmr(time, ncol)float647.99e-10 7.99e-10 ... 7.991e-10
- long_name :
- f11 volume mixing ratio
array([[7.99042148e-10, 7.99042148e-10, 7.99042148e-10], [7.99043693e-10, 7.99043693e-10, 7.99043693e-10], [7.99045238e-10, 7.99045238e-10, 7.99045238e-10], [7.99046783e-10, 7.99046783e-10, 7.99046783e-10], [7.99048329e-10, 7.99048329e-10, 7.99048329e-10], [7.99049874e-10, 7.99049874e-10, 7.99049874e-10], [7.99051419e-10, 7.99051419e-10, 7.99051419e-10], [7.99052964e-10, 7.99052964e-10, 7.99052964e-10], [7.99054510e-10, 7.99054510e-10, 7.99054510e-10], [7.99056055e-10, 7.99056055e-10, 7.99056055e-10], [7.99057600e-10, 7.99057600e-10, 7.99057600e-10], [7.99059145e-10, 7.99059145e-10, 7.99059145e-10], [7.99060691e-10, 7.99060691e-10, 7.99060691e-10], [7.99062236e-10, 7.99062236e-10, 7.99062236e-10], [7.99063781e-10, 7.99063781e-10, 7.99063781e-10], [7.99065327e-10, 7.99065327e-10, 7.99065327e-10], [7.99066872e-10, 7.99066872e-10, 7.99066872e-10], [7.99068417e-10, 7.99068417e-10, 7.99068417e-10], [7.99069962e-10, 7.99069962e-10, 7.99069962e-10], [7.99071508e-10, 7.99071508e-10, 7.99071508e-10], [7.99073053e-10, 7.99073053e-10, 7.99073053e-10], [7.99074598e-10, 7.99074598e-10, 7.99074598e-10], [7.99076143e-10, 7.99076143e-10, 7.99076143e-10], [7.99077689e-10, 7.99077689e-10, 7.99077689e-10]]) - f12vmr(time, ncol)float645.236e-10 5.236e-10 ... 5.236e-10
- long_name :
- f12 volume mixing ratio
array([[5.23569087e-10, 5.23569087e-10, 5.23569087e-10], [5.23568777e-10, 5.23568777e-10, 5.23568777e-10], [5.23568466e-10, 5.23568466e-10, 5.23568466e-10], [5.23568156e-10, 5.23568156e-10, 5.23568156e-10], [5.23567846e-10, 5.23567846e-10, 5.23567846e-10], [5.23567535e-10, 5.23567535e-10, 5.23567535e-10], [5.23567225e-10, 5.23567225e-10, 5.23567225e-10], [5.23566914e-10, 5.23566914e-10, 5.23566914e-10], [5.23566604e-10, 5.23566604e-10, 5.23566604e-10], [5.23566294e-10, 5.23566294e-10, 5.23566294e-10], [5.23565983e-10, 5.23565983e-10, 5.23565983e-10], [5.23565673e-10, 5.23565673e-10, 5.23565673e-10], [5.23565362e-10, 5.23565362e-10, 5.23565362e-10], [5.23565052e-10, 5.23565052e-10, 5.23565052e-10], [5.23564742e-10, 5.23564742e-10, 5.23564742e-10], [5.23564431e-10, 5.23564431e-10, 5.23564431e-10], [5.23564121e-10, 5.23564121e-10, 5.23564121e-10], [5.23563811e-10, 5.23563811e-10, 5.23563811e-10], [5.23563500e-10, 5.23563500e-10, 5.23563500e-10], [5.23563190e-10, 5.23563190e-10, 5.23563190e-10], [5.23562879e-10, 5.23562879e-10, 5.23562879e-10], [5.23562569e-10, 5.23562569e-10, 5.23562569e-10], [5.23562259e-10, 5.23562259e-10, 5.23562259e-10], [5.23561948e-10, 5.23561948e-10, 5.23561948e-10]]) - sol_tsi(time, ncol)float64-1.0 -1.0 -1.0 ... -1.0 -1.0 -1.0
- long_name :
- total solar irradiance
- units :
- W/m2
array([[-1., -1., -1.], [-1., -1., -1.], [-1., -1., -1.], [-1., -1., -1.], [-1., -1., -1.], [-1., -1., -1.], [-1., -1., -1.], [-1., -1., -1.], [-1., -1., -1.], [-1., -1., -1.], [-1., -1., -1.], [-1., -1., -1.], [-1., -1., -1.], [-1., -1., -1.], [-1., -1., -1.], [-1., -1., -1.], [-1., -1., -1.], [-1., -1., -1.], [-1., -1., -1.], [-1., -1., -1.], [-1., -1., -1.], [-1., -1., -1.], [-1., -1., -1.], [-1., -1., -1.]]) - nsteph(time, ncol)int3232114 32114 32114 ... 32160 32160
- long_name :
- current timestep
array([[32114, 32114, 32114], [32116, 32116, 32116], [32118, 32118, 32118], [32120, 32120, 32120], [32122, 32122, 32122], [32124, 32124, 32124], [32126, 32126, 32126], [32128, 32128, 32128], [32130, 32130, 32130], [32132, 32132, 32132], [32134, 32134, 32134], [32136, 32136, 32136], [32138, 32138, 32138], [32140, 32140, 32140], [32142, 32142, 32142], [32144, 32144, 32144], [32146, 32146, 32146], [32148, 32148, 32148], [32150, 32150, 32150], [32152, 32152, 32152], [32154, 32154, 32154], [32156, 32156, 32156], [32158, 32158, 32158], [32160, 32160, 32160]], dtype=int32) - ACTNI(time, ncol)float320.0 0.0 0.0 ... 2.371e+04 2.226e+04
- units :
- Micron
- long_name :
- Average Cloud Top ice number
- basename :
- ACTNI
array([[ 0. , 0. , 0. ], [ 8557.338 , 0. , 0. ], [ 2185.564 , 0. , 0. ], [ 3831.4524, 0. , 0. ], [ 3190.785 , 0. , 0. ], [ 8720.562 , 0. , 0. ], [ 20715.457 , 9476.448 , 3041.7554], [ 31962.346 , 6461.9336, 4046.7056], [ 34525.113 , 15306.996 , 9314.903 ], [ 28424.334 , 17412.85 , 9755.762 ], [ 18736.227 , 22751.783 , 13881.63 ], [ 10703.406 , 21750.72 , 18234.8 ], [ 3216.291 , 17995.281 , 16531.074 ], [ 1267.0157, 11974.873 , 12241.5625], [758922.8 , 5013.126 , 5331.2173], [ 19471.78 , 6522.024 , 1798.7648], [ 74358.58 , 16087.712 , 8368.193 ], [ 14553.429 , 14132.614 , 12102.414 ], [ 16908.633 , 8198.062 , 6487.5327], [ 18609.71 , 5317.317 , 3521.7761], [ 19512.51 , 4584.3867, 1490.0862], [ 3814.3347, 21297.027 , 20083.363 ], [ 41766.78 , 22866.082 , 21560.393 ], [ 44053.45 , 23709.127 , 22263.83 ]], dtype=float32) - ACTNL(time, ncol)float327.74e+05 7.677e+05 ... 0.0 0.0
- units :
- Micron
- long_name :
- Average Cloud Top droplet number
- basename :
- ACTNL
array([[7.7397106e+05, 7.6769325e+05, 6.3572670e+06], [0.0000000e+00, 1.5141961e+05, 1.1215826e+03], [0.0000000e+00, 3.9255648e+06, 6.7115060e+06], [0.0000000e+00, 2.7234353e+05, 7.0432875e+06], [0.0000000e+00, 2.4377195e+06, 7.9776145e+06], [0.0000000e+00, 7.5393845e+01, 3.3006285e+06], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], dtype=float32) - ACTREI(time, ncol)float320.0 0.0 0.0 ... 71.42 44.88 46.68
- units :
- Micron
- long_name :
- Average Cloud Top ice effective radius
- basename :
- ACTREI
array([[ 0. , 0. , 0. ], [29.185854, 0. , 0. ], [37.942253, 0. , 0. ], [40.049194, 0. , 0. ], [42.136127, 0. , 0. ], [40.377293, 0. , 0. ], [34.924427, 20.825615, 14.015091], [28.507236, 30.458511, 24.189386], [32.063515, 19.612564, 17.150776], [35.301277, 16.961077, 14.527979], [38.56587 , 20.120193, 16.700354], [40.704823, 29.697645, 25.212013], [42.44824 , 35.677147, 34.318676], [50.109352, 38.418274, 36.664425], [27.842411, 41.797993, 40.649006], [18.63569 , 43.067215, 43.85847 ], [37.700462, 39.503475, 41.8404 ], [50.358826, 35.682705, 37.789185], [49.45762 , 27.857752, 33.82564 ], [46.48117 , 18.007597, 24.24863 ], [40.129593, 46.73848 , 48.267353], [15.087814, 47.71531 , 48.236847], [50.0916 , 46.88475 , 47.694187], [71.42062 , 44.88017 , 46.677193]], dtype=float32) - ACTREL(time, ncol)float329.533 20.93 1.101 ... 0.0 0.0 0.0
- units :
- Micron
- long_name :
- Average Cloud Top droplet effective radius
- basename :
- ACTREL
array([[ 9.533483 , 20.934862 , 1.1009611 ], [ 0. , 28.352602 , 1.4560286 ], [ 0. , 7.6563544 , 17.115618 ], [ 0. , 6.0080304 , 15.354406 ], [ 0. , 20.747929 , 12.666198 ], [ 0. , 0.52703583, 14.597877 ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ]], dtype=float32) - ADRAIN(time, lev, ncol)float3255.72 13.97 12.86 ... nan nan nan
- mdims :
- 9
- units :
- Micron
- long_name :
- Average rain effective Diameter
- basename :
- ADRAIN
array([[[55.72431 , 13.965531, 12.863635], [42.64484 , 10. , 13.449966], [29.769634, 10. , 14.660041], ..., [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan]], [[54.873734, nan, 13.894699], [41.86107 , 10.768498, 14.566078], [29.126762, 14.291217, 15.941736], ..., [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan]], [[45.75345 , 10. , 12.885951], [43.76058 , 11.717133, 14.372966], [33.783154, 14.394375, 17.123003], ..., ... ..., [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan]], [[52.41884 , 49.008247, nan], [43.97372 , 41.661095, nan], [33.87179 , 32.787727, nan], ..., [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan]], [[50.06032 , 50.821552, 56.918602], [41.457058, 43.521545, 48.124565], [31.143415, 34.5488 , 37.298603], ..., [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan]]], dtype=float32) - ADSNOW(time, lev, ncol)float32nan 187.8 143.8 nan ... nan nan nan
- mdims :
- 9
- units :
- Micron
- long_name :
- Average snow effective Diameter
- basename :
- ADSNOW
array([[[ nan, 187.8456 , 143.80016], [ nan, 186.9749 , 131.08989], [ nan, 169.75093, 129.291 ], ..., [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan]], [[ nan, 190.20515, 146.30928], [ nan, 189.92955, 133.4125 ], [ nan, 173.35652, 131.67088], ..., [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan]], [[ nan, 188.16035, 141.89883], [ nan, 171.9416 , 129.55003], [ nan, 170.85492, 128.47256], ..., ... ..., [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan]], [[ 428.5892 , 379.9916 , 1223.0029 ], [ 402.13043, 362.0272 , 1027.803 ], [ 362.50583, 335.31177, 792.3841 ], ..., [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan]], [[ 404.29175, 353.80515, 437.46613], [ 382.61404, 337.3432 , 403.1429 ], [ 350.1658 , 312.9682 , 354.79236], ..., [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan]]], dtype=float32) - ANRAIN(time, lev, ncol)float320.7012 0.6562 2.89 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- m-3
- long_name :
- Average rain number conc
- basename :
- ANRAIN
array([[[7.0116162e-01, 6.5618539e-01, 2.8896465e+00], [1.0301039e+00, 8.4216917e-01, 3.3427464e+01], [3.5078678e+00, 8.5738678e+01, 8.3302600e+02], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[5.7783538e-01, 5.8280432e-01, 3.4371777e+00], [8.5047930e-01, 1.0660580e+00, 4.2543900e+01], [2.9005358e+00, 3.1034061e+02, 1.1128694e+03], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[1.5508625e-01, 1.2410737e+00, 5.8738966e+00], [3.7922254e-01, 4.2767227e+01, 9.0603447e+01], [1.1252331e+00, 8.8724799e+02, 1.8747097e+03], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[8.1586218e-01, 9.7617340e-01, 0.0000000e+00], [1.4884335e+00, 2.9634106e+00, 0.0000000e+00], [8.5535316e+00, 1.7280516e+01, 6.7465240e-03], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[8.4381670e-01, 6.5035909e-01, 8.6978510e-02], [1.6008216e+00, 1.9903331e+00, 1.3844140e-01], [1.0518725e+01, 1.1669583e+01, 2.9919252e-01], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - ANSNOW(time, lev, ncol)float320.0 0.00412 0.1215 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- m-3
- long_name :
- Average snow number conc
- basename :
- ANSNOW
array([[[0.0000000e+00, 4.1197925e-03, 1.2154137e-01], [0.0000000e+00, 5.1872875e-03, 1.4697193e+00], [0.0000000e+00, 3.2101676e-01, 2.1330063e+01], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 4.3584830e-03, 1.4223434e-01], [0.0000000e+00, 5.5329432e-03, 1.9335165e+00], [0.0000000e+00, 9.8544782e-01, 2.8241529e+01], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 4.5465897e-03, 2.6838461e-01], [0.0000000e+00, 1.1072798e-01, 3.9824915e+00], [0.0000000e+00, 1.6831764e+00, 5.1641216e+01], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[2.3968600e-02, 8.3912209e-02, 1.1182872e-03], [3.2775246e-02, 1.9389229e-01, 1.5630857e-03], [1.3525411e-01, 7.6747781e-01, 2.7517087e-03], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[2.0404946e-02, 5.6029070e-02, 1.6321024e-02], [2.7254228e-02, 1.3150753e-01, 1.8906288e-02], [1.1103019e-01, 5.3080767e-01, 2.4763001e-02], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - AODVIS(time, ncol)float320.03071 0.03278 ... 0.09031 0.08787
- long_name :
- Aerosol optical depth 550 nm
- basename :
- AODVIS
array([[0.03071209, 0.03277841, 0.03339631], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, 0.02891912], [0.11428536, 0.04497121, 0.03348441], [0.09706142, 0.0633003 , 0.04073798], [0.09128533, 0.06452267, 0.05115647], [0.10922865, 0.09278391, 0.07076322], [0.12893279, 0.09030766, 0.08787203]], dtype=float32) - AQRAIN(time, lev, ncol)float321.48e-10 2.503e-11 ... 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Average rain mixing ratio
- basename :
- AQRAIN
array([[[1.4796600e-10, 2.5025435e-11, 1.4693149e-10], [1.9144121e-10, 4.1799782e-11, 1.9501698e-09], [5.6471416e-10, 5.5677338e-09, 5.9480517e-08], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[1.1400540e-10, 2.2607300e-11, 2.2786947e-10], [1.4737818e-10, 4.6271955e-11, 3.1467542e-09], [4.3401210e-10, 1.9020069e-08, 9.9270387e-08], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[1.9047700e-10, 8.4802262e-11, 3.8851561e-10], [2.4701019e-10, 3.7159458e-09, 7.1077531e-09], [3.5778944e-10, 1.1105296e-07, 1.9777643e-07], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[9.3734964e-10, 9.3642705e-10, 0.0000000e+00], [1.2292618e-09, 2.1359601e-09, 0.0000000e+00], [4.4596447e-09, 8.1062836e-09, 1.3524976e-11], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[9.3274855e-10, 7.3832951e-10, 1.7780903e-10], [1.2330386e-09, 1.7084596e-09, 1.9897498e-10], [4.7888524e-09, 6.5883117e-09, 2.4917188e-10], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - AQSNOW(time, lev, ncol)float320.0 1.252e-10 1.558e-09 ... 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Average snow mixing ratio
- basename :
- AQSNOW
array([[[0.0000000e+00, 1.2515441e-10, 1.5580778e-09], [0.0000000e+00, 1.4367420e-10, 1.6159730e-08], [0.0000000e+00, 8.4039931e-09, 2.2470799e-07], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 1.3520318e-10, 1.9641186e-09], [0.0000000e+00, 1.5757826e-10, 2.2719576e-08], [0.0000000e+00, 2.6870014e-08, 3.1764463e-07], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 1.2316292e-10, 3.4538652e-09], [0.0000000e+00, 2.9710998e-09, 4.3623434e-08], [0.0000000e+00, 4.3328733e-08, 5.4764951e-07], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[6.6547936e-09, 1.9569995e-08, 5.4650018e-09], [8.1822789e-09, 4.1933038e-08, 5.4773408e-09], [2.7638604e-08, 1.4267818e-07, 5.5014517e-09], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[4.8890194e-09, 9.9272759e-09, 5.7880962e-09], [5.9829155e-09, 2.1675502e-08, 5.7902074e-09], [2.0662629e-08, 7.5720649e-08, 5.7977103e-09], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - AREI(time, lev, ncol)float32nan nan nan nan ... nan nan nan nan
- mdims :
- 9
- units :
- Micron
- long_name :
- Average ice effective radius
- basename :
- AREI
array([[[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., ... ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]]], dtype=float32) - AREL(time, lev, ncol)float32nan nan nan nan ... nan nan nan nan
- mdims :
- 9
- units :
- Micron
- long_name :
- Average droplet effective radius
- basename :
- AREL
array([[[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., ... ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]]], dtype=float32) - AWNC(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- m-3
- long_name :
- Average cloud water number conc
- basename :
- AWNC
array([[[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., ... ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - AWNI(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- m-3
- long_name :
- Average cloud ice number conc
- basename :
- AWNI
array([[[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., ... ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - CCN3(time, lev, ncol)float3210.09 9.887 10.95 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- #/cm3
- long_name :
- CCN concentration at S=0.1%
- basename :
- CCN3
array([[[10.087448 , 9.887123 , 10.952791 ], [10.027431 , 9.827665 , 10.880567 ], [ 9.931998 , 9.732036 , 10.749885 ], ..., [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ]], [[10.045017 , 9.539835 , 10.676319 ], [ 9.983613 , 9.4829445, 10.61025 ], [ 9.885693 , 9.391727 , 10.491909 ], ..., [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ]], [[10.231188 , 9.172615 , 10.389794 ], [10.16933 , 9.117474 , 10.329419 ], [10.07049 , 9.028815 , 10.22403 ], ..., ... ..., [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ]], [[18.690313 , 18.332838 , 13.983971 ], [18.529238 , 18.205244 , 13.857922 ], [18.230757 , 17.984264 , 13.595864 ], ..., [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ]], [[17.697273 , 18.781399 , 13.140232 ], [17.542122 , 18.641277 , 12.998683 ], [17.254387 , 18.390368 , 12.686504 ], ..., [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ]]], dtype=float32) - CDNUMC(time, ncol)float321.169e+10 1.565e+10 ... 2.56e+10
- units :
- 1/m2
- long_name :
- Vertically-integrated droplet concentration
- basename :
- CDNUMC
array([[1.16894925e+10, 1.56514345e+10, 1.60180675e+10], [1.18583480e+10, 1.41108398e+10, 1.40021176e+10], [1.22299177e+10, 1.44471398e+10, 1.34825359e+10], [1.30057165e+10, 1.32761242e+10, 1.33348004e+10], [1.32274954e+10, 1.27735194e+10, 1.30376745e+10], [1.36546929e+10, 1.34120069e+10, 1.13390090e+10], [1.51058790e+10, 1.40114596e+10, 1.20241910e+10], [1.60714066e+10, 1.43367854e+10, 1.17917030e+10], [1.43830170e+10, 1.35338435e+10, 9.89781606e+09], [1.71680420e+10, 1.20526592e+10, 9.65850829e+09], [3.11448392e+10, 9.94922906e+09, 9.21176883e+09], [2.93855621e+10, 7.20925184e+09, 4.74477978e+09], [4.36743782e+10, 6.07978957e+09, 1.23341102e-03], [4.23258440e+10, 5.94076058e+09, 1.04539755e-08], [4.46905426e+10, 3.66662016e+09, 1.04502860e-08], [4.58525245e+10, 1.13817221e+10, 1.04450812e-08], [3.97381181e+10, 2.57466061e+10, 4.33923533e+09], [1.79018936e+10, 2.43351368e+10, 1.77391985e+10], [1.34875515e+10, 1.86821530e+10, 1.27409848e+10], [1.44694313e+10, 1.50782925e+10, 1.20307149e+10], [3.34416916e+10, 1.99374500e+10, 1.27197481e+10], [3.43862845e+10, 3.33380321e+10, 2.19615846e+10], [2.88043909e+10, 3.07499622e+10, 3.62409124e+10], [3.06624471e+10, 2.28480512e+10, 2.56025784e+10]], dtype=float32) - CLDHGH(time, ncol)float320.00104 0.0 0.0 ... 0.0 0.999 0.999
- units :
- fraction
- long_name :
- Vertically-integrated high cloud
- basename :
- CLDHGH
array([[0.00103972, 0. , 0. ], [0.8083792 , 0. , 0. ], [0.999 , 0. , 0. ], [0.999 , 0.00335687, 0. ], [0.999 , 0.14378357, 0.04888189], [0.999 , 0.41002554, 0.19591814], [0.999 , 0.8907955 , 0.508126 ], [0.999 , 0.88918316, 0.75176764], [0.999 , 0.6935814 , 0.62590224], [0.999 , 0.63216287, 0.560871 ], [0.999 , 0.7494317 , 0.6310767 ], [0.999 , 0.91709584, 0.8227196 ], [0.999 , 0.999 , 0.99823326], [0.999 , 0.999 , 0.999 ], [0.999 , 0.999 , 0.999 ], [0.999 , 0.999 , 0.999 ], [0.999 , 0.999 , 0.999 ], [0.999 , 0.999 , 0.999 ], [0.999 , 0.999 , 0.999 ], [0.999 , 0.999 , 0.999 ], [0.999 , 0.999 , 0.999 ], [0.8093421 , 0.999 , 0.999 ], [0.07882405, 0.999 , 0.999 ], [0. , 0.999 , 0.999 ]], dtype=float32) - CLDICE(time, lev, ncol)float324.923e-15 1.407e-14 ... 3.773e-24
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Grid box averaged cloud ice amount
- basename :
- CLDICE
array([[[4.9226556e-15, 1.4069195e-14, 3.3578623e-16], [1.9024746e-13, 8.9400999e-15, 6.9678405e-13], [8.1440887e-12, 4.4209743e-14, 7.8421897e-10], ..., [4.2775994e-24, 4.2770212e-24, 4.2428320e-24], [3.8617147e-24, 3.7071402e-24, 3.7020225e-24], [3.6965450e-24, 3.6641903e-24, 3.6970625e-24]], [[2.9488481e-16, 1.2808379e-14, 4.1564693e-16], [1.5516187e-14, 7.8176703e-15, 2.2599325e-12], [9.2084413e-13, 5.1194922e-14, 2.0499316e-09], ..., [4.2435226e-24, 4.2724363e-24, 4.2524044e-24], [3.8657458e-24, 3.7466519e-24, 3.7543421e-24], [3.7516308e-24, 3.6766231e-24, 3.6929364e-24]], [[8.2111417e-13, 9.6426284e-16, 7.3736399e-16], [7.5937802e-13, 1.0103278e-14, 9.6290918e-12], [8.6103596e-13, 5.7849233e-14, 6.7653696e-09], ..., ... ..., [4.3507612e-24, 4.2892198e-24, 4.2676480e-24], [3.9637898e-24, 3.9489355e-24, 3.9434167e-24], [3.6608917e-24, 3.6868471e-24, 3.6891459e-24]], [[9.9630312e-17, 3.0062036e-16, 2.6899510e-14], [1.1806295e-16, 6.3765583e-16, 2.4054153e-14], [3.2260986e-16, 9.0063520e-13, 9.1016053e-15], ..., [4.3660828e-24, 4.3232548e-24, 4.2977742e-24], [3.9835444e-24, 3.9754208e-24, 3.9374107e-24], [3.7604849e-24, 3.7288161e-24, 3.7131170e-24]], [[9.8204246e-17, 1.1110487e-16, 5.5393759e-14], [1.1426775e-16, 7.2294590e-16, 3.7547788e-14], [3.2725973e-16, 1.6132430e-14, 1.1006347e-14], ..., [4.3689085e-24, 4.3351902e-24, 4.3099759e-24], [4.0027071e-24, 3.9561225e-24, 3.8853735e-24], [3.8223959e-24, 3.7787021e-24, 3.7734672e-24]]], dtype=float32) - CLDLIQ(time, lev, ncol)float324.305e-36 0.0 0.0 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Grid box averaged cloud liquid amount
- basename :
- CLDLIQ
array([[[4.3050988e-36, 0.0000000e+00, 0.0000000e+00], [1.9900146e-35, 0.0000000e+00, 0.0000000e+00], [5.9071848e-22, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[8.2156167e-38, 0.0000000e+00, 0.0000000e+00], [8.8877467e-38, 0.0000000e+00, 0.0000000e+00], [6.4625698e-31, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [1.2640355e-34, 0.0000000e+00, 0.0000000e+00], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [4.5011802e-18, 4.3241853e-17, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [1.7005988e-17, 1.8569171e-16, 2.3869881e-18], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - CLDLOW(time, ncol)float321.0 1.0 0.9999 1.0 ... 1.0 1.0 1.0
- units :
- fraction
- long_name :
- Vertically-integrated low cloud
- basename :
- CLDLOW
array([[1.0000000e+00, 9.9998808e-01, 9.9990749e-01], [9.9999994e-01, 9.9997175e-01, 9.9989533e-01], [1.0000000e+00, 9.9994689e-01, 9.9994057e-01], [1.0000000e+00, 9.9991578e-01, 1.0000000e+00], [1.0000000e+00, 9.9987549e-01, 9.9993908e-01], [1.0000000e+00, 9.9990386e-01, 9.9991709e-01], [9.9999994e-01, 9.9995375e-01, 9.9994344e-01], [1.0000000e+00, 9.9991465e-01, 9.9973869e-01], [9.9999267e-01, 9.9981546e-01, 9.9978632e-01], [9.9999654e-01, 9.9969631e-01, 1.0000000e+00], [1.0000000e+00, 9.9900001e-01, 1.0000000e+00], [9.9999332e-01, 9.9900001e-01, 1.0000000e+00], [1.0000000e+00, 1.0000000e+00, 9.9900001e-01], [9.9999058e-01, 1.0000000e+00, 9.9900001e-01], [1.0000000e+00, 6.9922006e-01, 4.9262837e-02], [1.0000000e+00, 1.0000000e+00, 0.0000000e+00], [1.0000000e+00, 1.0000000e+00, 0.0000000e+00], [1.0000000e+00, 1.0000000e+00, 1.3397803e-05], [1.0000000e+00, 8.9895475e-01, 5.8531690e-01], [1.0000000e+00, 9.9942338e-01, 9.9900001e-01], [1.0000000e+00, 1.0000000e+00, 1.0000000e+00], [1.0000000e+00, 1.0000000e+00, 1.0000000e+00], [1.0000000e+00, 1.0000000e+00, 1.0000000e+00], [1.0000000e+00, 1.0000000e+00, 1.0000000e+00]], dtype=float32) - CLDMED(time, ncol)float320.0 0.0 0.0 0.0 ... 1.0 1.0 1.0 1.0
- units :
- fraction
- long_name :
- Vertically-integrated mid-level cloud
- basename :
- CLDMED
array([[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [1.3352673e-01, 0.0000000e+00, 0.0000000e+00], [5.7944614e-01, 0.0000000e+00, 0.0000000e+00], [9.9900001e-01, 0.0000000e+00, 0.0000000e+00], [9.9900001e-01, 0.0000000e+00, 0.0000000e+00], [9.9900001e-01, 0.0000000e+00, 0.0000000e+00], [9.9900001e-01, 8.4039703e-02, 5.0260314e-05], [9.9900001e-01, 5.1391071e-01, 1.3083148e-01], [1.0000000e+00, 9.9900001e-01, 5.5851430e-01], [1.0000000e+00, 9.9900001e-01, 9.9900001e-01], [1.0000000e+00, 9.9900001e-01, 9.9900001e-01], [1.0000000e+00, 9.9900001e-01, 9.9900001e-01], [1.0000000e+00, 9.9900001e-01, 9.9900001e-01], [1.0000000e+00, 1.0000000e+00, 9.9900001e-01], [1.0000000e+00, 1.0000000e+00, 9.9900001e-01], [1.0000000e+00, 1.0000000e+00, 1.0000000e+00], [1.0000000e+00, 1.0000000e+00, 9.9997431e-01], [1.0000000e+00, 1.0000000e+00, 1.0000000e+00], [1.0000000e+00, 1.0000000e+00, 1.0000000e+00], [1.0000000e+00, 1.0000000e+00, 1.0000000e+00], [1.0000000e+00, 1.0000000e+00, 1.0000000e+00], [1.0000000e+00, 1.0000000e+00, 1.0000000e+00]], dtype=float32) - CLDTOT(time, ncol)float321.0 1.0 0.9999 1.0 ... 1.0 1.0 1.0
- units :
- fraction
- long_name :
- Vertically-integrated total cloud
- basename :
- CLDTOT
array([[1. , 0.9999881 , 0.9999075 ], [0.99999994, 0.99997175, 0.99989533], [1. , 0.9999469 , 0.9999406 ], [1. , 0.9999158 , 1. ], [1. , 0.9998755 , 0.9999391 ], [1. , 0.99990386, 0.9999171 ], [0.99999994, 0.99995375, 0.99994344], [1. , 0.99991465, 0.9997387 ], [0.99999267, 0.99981546, 0.9997863 ], [0.99999654, 0.9996963 , 1. ], [1. , 0.999 , 1. ], [1. , 0.999 , 1. ], [1. , 1. , 0.999 ], [1. , 1. , 0.999 ], [1. , 0.999 , 0.999 ], [1. , 1. , 0.999 ], [1. , 1. , 0.999 ], [1. , 1. , 1. ], [1. , 1. , 0.9999743 ], [1. , 1. , 1. ], [1. , 1. , 1. ], [1. , 1. , 1. ], [1. , 1. , 1. ], [1. , 1. , 1. ]], dtype=float32) - CLOUD(time, lev, ncol)float322.374e-05 0.0001 ... 0.0 0.0
- mdims :
- 9
- units :
- fraction
- long_name :
- Cloud fraction
- basename :
- CLOUD
array([[[2.3736337e-05, 9.9999997e-05, 5.1592407e-04], [6.2959516e-05, 9.9999997e-05, 3.8299891e-03], [1.4467775e-04, 5.2718641e-03, 4.5738880e-02], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[2.2758020e-05, 9.9999997e-05, 6.3581247e-04], [6.4255721e-05, 9.9999997e-05, 5.1842085e-03], [1.4255832e-04, 1.5475810e-02, 6.3984610e-02], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 9.9999997e-05, 1.1370420e-03], [1.6439892e-05, 2.3446756e-03, 9.8446077e-03], [4.7910871e-05, 3.0964246e-02, 1.1348072e-01], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[5.5862369e-05, 3.0046186e-04, 0.0000000e+00], [1.2313537e-04, 6.4018444e-04, 0.0000000e+00], [4.1422047e-04, 2.1369918e-03, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[5.4536562e-05, 1.1765216e-04, 0.0000000e+00], [1.2218198e-04, 2.5570864e-04, 0.0000000e+00], [4.1664098e-04, 8.7710691e-04, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - CMFDICE(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg/s
- long_name :
- Cloud ice tendency - shallow convection
- basename :
- CMFDICE
array([[[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., ... ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - CMFDLIQ(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg/s
- long_name :
- Cloud liq tendency - shallow convection
- basename :
- CMFDLIQ
array([[[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., ... ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - CMFDQ(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg/s
- long_name :
- QV tendency - shallow convection
- basename :
- CMFDQ
array([[[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., ... ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - CMFDT(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- K/s
- long_name :
- T tendency - shallow convection
- basename :
- CMFDT
array([[[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., ... ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - CONCLD(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- fraction
- long_name :
- Convective cloud cover
- basename :
- CONCLD
array([[[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., ... ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - EMIS(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- 1
- long_name :
- Cloud longwave emissivity
- basename :
- EMIS
array([[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 1.5230246e-05], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 2.9315594e-05], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 5.4004481e-09, 5.2262672e-05], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - EVAPPREC(time, lev, ncol)float324.923e-09 0.0 3.932e-09 ... 0.0 0.0
- mdims :
- 9
- units :
- kg/kg/s
- long_name :
- Rate of evaporation of falling precip
- basename :
- EVAPPREC
array([[[4.92341634e-09, 0.00000000e+00, 3.93212707e-09], [6.37081854e-09, 0.00000000e+00, 6.06938722e-09], [1.29959918e-08, 0.00000000e+00, 8.42343173e-09], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[3.79439591e-09, 0.00000000e+00, 3.84389276e-09], [4.90591656e-09, 0.00000000e+00, 6.09981710e-09], [1.01390505e-08, 5.67312597e-09, 8.80847661e-09], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[5.91090288e-09, 0.00000000e+00, 3.81477649e-09], [8.22080004e-09, 6.40319486e-09, 6.41847198e-09], [1.19149490e-08, 1.31095890e-08, 9.03894204e-09], ..., ... ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[7.37419104e-09, 3.15597282e-09, 1.85027701e-10], [8.50655635e-09, 3.64512931e-09, 1.47808349e-10], [8.00337929e-09, 4.30754543e-09, 5.21626180e-11], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[7.66214203e-09, 3.42936546e-09, 7.99513122e-10], [9.43340250e-09, 3.56113916e-09, 5.81052872e-10], [9.56001145e-09, 3.41530138e-09, 1.80768220e-10], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]]], dtype=float32) - EVAPSNOW(time, lev, ncol)float324.803e-13 0.0 3.323e-09 ... 0.0 0.0
- mdims :
- 9
- units :
- kg/kg/s
- long_name :
- Rate of evaporation of falling snow
- basename :
- EVAPSNOW
array([[[4.8033084e-13, 0.0000000e+00, 3.3226906e-09], [3.8311742e-13, 0.0000000e+00, 4.5781960e-09], [2.4037814e-13, 0.0000000e+00, 4.1756665e-09], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[3.9146580e-13, 0.0000000e+00, 3.1025911e-09], [3.1312838e-13, 0.0000000e+00, 4.3276303e-09], [1.9882952e-13, 1.6089178e-09, 3.7386121e-09], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[3.0213123e-13, 0.0000000e+00, 2.9802838e-09], [2.4635526e-13, 1.3648693e-09, 4.2100883e-09], [1.6122047e-13, 1.2430513e-09, 3.3181577e-09], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[2.8321578e-09, 2.9861409e-09, 1.8313948e-10], [2.7704603e-09, 3.3112386e-09, 1.4540184e-10], [3.8392054e-09, 3.3761662e-09, 4.8753685e-11], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[2.5286555e-09, 2.6627158e-09, 7.6169632e-10], [2.4353926e-09, 2.9191338e-09, 5.2378046e-10], [3.3174634e-09, 2.8638405e-09, 7.0503305e-11], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - FCTI(time, ncol)float320.0 0.0 0.0 ... 0.999 0.999 0.999
- units :
- fraction
- long_name :
- Fractional occurrence of cloud top ice
- basename :
- FCTI
array([[0. , 0. , 0. ], [0.8083792 , 0. , 0. ], [0.999 , 0. , 0. ], [0.999 , 0. , 0. ], [0.999 , 0. , 0. ], [0.999 , 0. , 0. ], [0.999 , 0.67876786, 0.508126 ], [0.89592344, 0.88918316, 0.75176764], [0.977181 , 0.6935814 , 0.62590224], [0.999 , 0.63216287, 0.560871 ], [0.999 , 0.72118723, 0.6266658 ], [0.999 , 0.91709584, 0.8227196 ], [0.999 , 0.999 , 0.99823326], [0.999 , 0.999 , 0.999 ], [0.999 , 0.999 , 0.999 ], [0.66232 , 0.999 , 0.999 ], [0.999 , 0.999 , 0.999 ], [0.999 , 0.999 , 0.999 ], [0.999 , 0.8459101 , 0.9638587 ], [0.999 , 0.6611711 , 0.77635694], [0.9977829 , 0.999 , 0.999 ], [0.5285695 , 0.999 , 0.999 ], [0.999 , 0.999 , 0.999 ], [0.999 , 0.999 , 0.999 ]], dtype=float32) - FCTL(time, ncol)float320.4317 0.999 0.189 ... 0.0 0.0 0.0
- units :
- fraction
- long_name :
- Fractional occurrence of cloud top liquid
- basename :
- FCTL
array([[0.43168664, 0.999 , 0.18904454], [0. , 0.999 , 0.0507626 ], [0. , 0.999 , 0.999 ], [0. , 0.7710168 , 1. ], [0. , 0.999 , 0.999 ], [0. , 0.01837454, 0.999 ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ]], dtype=float32) - FDL(time, ncol, ilev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 10
- units :
- W/m2
- long_name :
- Longwave downward flux
- basename :
- FDL
array([[[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], [[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], [[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], ..., [[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], [[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], [[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]]], dtype=float32) - FDLC(time, ncol, ilev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 10
- units :
- W/m2
- long_name :
- Longwave clear-sky downward flux
- basename :
- FDLC
array([[[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], [[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], [[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], ..., [[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], [[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], [[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]]], dtype=float32) - FDS(time, ncol, ilev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 10
- units :
- W/m2
- long_name :
- Shortwave downward flux
- basename :
- FDS
array([[[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], [[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], [[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], ..., [[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], [[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], [[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]]], dtype=float32) - FDSC(time, ncol, ilev)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 10
- units :
- W/m2
- long_name :
- Shortwave clear-sky downward flux
- basename :
- FDSC
array([[[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], [[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], [[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], ..., [[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], [[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]], [[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]]], dtype=float32) - FICE(time, lev, ncol)float320.001445 0.8334 0.9138 ... 0.0 0.0
- mdims :
- 9
- units :
- fraction
- long_name :
- Fractional ice content within cloud
- basename :
- FICE
array([[[1.4454841e-03, 8.3336359e-01, 9.1382366e-01], [1.0339771e-03, 7.7463263e-01, 8.9231473e-01], [4.5222286e-04, 6.0149997e-01, 7.9072690e-01], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[1.5939100e-03, 8.5674399e-01, 8.9604437e-01], [1.1151755e-03, 7.7301002e-01, 8.7834561e-01], [4.5741317e-04, 5.8552986e-01, 7.6196784e-01], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[7.2889542e-04, 5.9222853e-01, 8.9888674e-01], [5.0388201e-04, 4.4430682e-01, 8.5989410e-01], [2.9065495e-04, 2.8065979e-01, 7.3495293e-01], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[8.7653691e-01, 9.5433491e-01, 9.9849594e-01], [8.6938781e-01, 9.5153147e-01, 9.9822980e-01], [8.6106271e-01, 9.4623923e-01, 9.9754757e-01], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[8.3978260e-01, 9.3077475e-01, 9.7019577e-01], [8.2912326e-01, 9.2693883e-01, 9.6677762e-01], [8.1184387e-01, 9.1995633e-01, 9.5879334e-01], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - FLDS(time, ncol)float32280.7 279.6 268.8 ... 294.2 283.4
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Downwelling longwave flux at surface
- basename :
- FLDS
array([[280.69238, 279.5848 , 268.81247], [280.73898, 280.55875, 269.60934], [280.73428, 281.1117 , 270.55765], [280.62527, 281.5819 , 270.97412], [280.52295, 282.0952 , 271.36642], [280.24716, 282.04788, 271.561 ], [280.0014 , 281.7211 , 271.31067], [279.72656, 281.2767 , 270.81897], [279.54318, 280.09525, 269.73972], [279.8138 , 278.20303, 268.45862], [280.28513, 276.70023, 264.67325], [281.11362, 272.76822, 242.96251], [282.88794, 259.34152, 225.07799], [285.0478 , 260.79016, 213.4504 ], [288.19315, 256.55664, 219.29738], [291.51593, 251.37383, 223.65433], [294.03168, 262.5557 , 233.09926], [296.73447, 261.82407, 249.48769], [299.87808, 270.3087 , 256.61465], [300.401 , 274.92416, 264.2902 ], [301.54843, 280.39655, 264.72757], [301.96033, 287.54373, 270.38773], [302.12268, 291.24042, 278.07443], [301.83682, 294.20407, 283.38895]], dtype=float32) - FLDSC(time, ncol)float32199.5 200.7 194.2 ... 217.8 208.6
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Clearsky Downwelling longwave flux at surface
- basename :
- FLDSC
array([[199.45505, 200.6656 , 194.24307], [199.69139, 201.43248, 194.76727], [199.83374, 202.09734, 195.29198], [200.00172, 202.71983, 195.91995], [200.0962 , 203.1552 , 196.39651], [200.31114, 203.44568, 196.76009], [200.65015, 203.58481, 196.96777], [201.28447, 203.5882 , 196.96017], [202.20958, 203.50243, 196.79395], [203.43958, 203.33086, 196.4611 ], [205.04779, 203.0933 , 196.06673], [207.01964, 202.8593 , 195.42273], [209.29767, 202.74529, 193.59747], [211.78474, 202.68143, 191.9114 ], [214.36734, 203.05722, 190.87985], [216.90399, 203.69518, 191.37105], [219.42882, 204.6364 , 192.31377], [221.62253, 205.93706, 193.70764], [223.50446, 207.32422, 195.67503], [225.09474, 209.00395, 197.58386], [226.09483, 210.97305, 199.87434], [226.7545 , 213.0335 , 202.43811], [227.0952 , 215.491 , 205.46333], [226.95184, 217.80408, 208.5933 ]], dtype=float32) - FLN200(time, ncol)float32181.6 175.8 171.9 ... 137.2 137.9
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Net longwave flux at 200 mb
- basename :
- FLN200
array([[181.63145, 175.78432, 171.92537], [181.46632, 176.63123, 172.33615], [180.69075, 177.01093, 172.84773], [179.61081, 177.35898, 173.32806], [178.48746, 177.6213 , 173.68597], [173.12837, 177.5207 , 174.12689], [165.3101 , 177.09372, 174.35172], [160.04568, 176.44109, 174.25577], [156.30577, 175.96399, 173.99644], [154.451 , 174.70705, 173.63492], [149.07567, 171.8142 , 173.09581], [144.46935, 167.69159, 174.99707], [142.95679, 165.44112, 170.55946], [140.55365, 161.28178, 165.46378], [140.0747 , 160.37535, 161.35063], [130.60341, 161.53316, 161.7308 ], [134.4604 , 142.10281, 150.46484], [133.2863 , 136.21169, 136.69405], [140.45877, 134.67688, 138.03511], [145.63287, 139.2056 , 142.445 ], [141.6708 , 142.80418, 144.25475], [135.6937 , 146.34378, 146.19223], [139.82283, 138.442 , 140.51271], [147.79951, 137.21866, 137.94554]], dtype=float32) - FLNS(time, ncol)float3233.26 28.08 14.88 ... 16.45 2.738
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Net longwave flux at surface
- basename :
- FLNS
array([[33.262386 , 28.076458 , 14.879767 ], [33.19853 , 27.268536 , 13.857829 ], [33.18599 , 26.927505 , 13.060074 ], [33.27775 , 26.659029 , 13.101119 ], [33.362812 , 26.308744 , 13.020462 ], [33.62132 , 26.465555 , 12.980641 ], [33.849827 , 26.815142 , 13.171859 ], [34.10743 , 27.19768 , 13.214188 ], [34.273537 , 28.23514 , 13.407933 ], [33.985653 , 29.84681 , 13.408607 ], [33.497086 , 30.969513 , 15.774937 ], [32.651333 , 34.653973 , 35.297207 ], [30.859768 , 47.881855 , 43.409813 ], [28.682705 , 45.608395 , 46.090366 ], [25.520098 , 50.369427 , 35.262463 ], [22.180082 , 55.572086 , 35.103825 ], [19.647112 , 44.417088 , 30.257277 ], [16.927076 , 45.848354 , 18.36789 ], [13.766231 , 37.49404 , 16.225924 ], [13.226058 , 33.354782 , 11.236076 ], [12.061398 , 28.291662 , 13.3824215], [11.632226 , 21.743204 , 9.650012 ], [11.452618 , 18.738386 , 4.758803 ], [11.721215 , 16.44939 , 2.7381837]], dtype=float32) - FLNT(time, ncol)float32200.9 195.6 192.1 ... 157.1 158.0
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Net longwave flux at top of model
- basename :
- FLNT
array([[200.90723, 195.60602, 192.05638], [200.71204, 196.39726, 192.42897], [199.9027 , 196.71967, 192.8777 ], [198.78297, 197.01344, 193.29637], [197.6929 , 197.25195, 193.62509], [192.50615, 197.1396 , 194.0176 ], [185.00961, 196.67488, 194.18846], [179.83078, 195.96284, 194.0043 ], [176.1282 , 195.41545, 193.65614], [174.2608 , 194.1051 , 193.21149], [168.96216, 191.24405, 192.58568], [164.48337, 187.25597, 194.25847], [162.92906, 184.89346, 189.98956], [160.5387 , 180.83022, 185.10435], [159.99771, 179.94891, 181.02621], [150.87563, 180.9767 , 181.29364], [154.43205, 162.226 , 170.38597], [153.26256, 156.63591, 157.17462], [160.05908, 155.03209, 158.38156], [164.94113, 159.28558, 162.58072], [161.00629, 162.63834, 164.23163], [155.25272, 165.94565, 165.99251], [159.16544, 158.2671 , 160.47804], [166.81964, 157.05074, 157.96744]], dtype=float32) - FLUT(time, ncol)float32201.2 195.9 192.4 ... 157.3 158.3
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Upwelling longwave flux at top of model
- basename :
- FLUT
array([[201.19269, 195.91656, 192.37566], [201.00763, 196.72093, 192.76154], [200.19627, 197.04715, 193.21114], [199.06737, 197.33434, 193.62299], [197.96565, 197.5612 , 193.94482], [192.76982, 197.43474, 194.3252 ], [185.26625, 196.96098, 194.48659], [180.08305, 196.24464, 194.29663], [176.3774 , 195.69421, 193.94502], [174.50429, 194.3774 , 193.49557], [169.19966, 191.50334, 192.8557 ], [164.71968, 187.50366, 194.51598], [163.166 , 185.13377, 190.23761], [160.7791 , 181.06902, 185.34995], [160.24316, 180.19089, 181.27469], [151.12617, 181.2226 , 181.54636], [154.68646, 162.4738 , 170.63956], [153.5171 , 156.88481, 157.42708], [160.31122, 155.28313, 158.63702], [165.19371, 159.54097, 162.84389], [161.2642 , 162.90495, 164.50737], [155.51704, 166.22649, 166.27759], [159.434 , 158.55186, 160.76585], [167.08875, 157.33684, 158.25694]], dtype=float32) - FLUTC(time, ncol)float32222.9 222.0 213.9 ... 218.6 210.6
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Clearsky upwelling longwave flux at top of model
- basename :
- FLUTC
array([[222.91275, 222.00468, 213.85579], [222.63602, 221.90807, 213.67801], [222.31342, 221.77852, 213.57503], [221.95903, 221.64247, 213.54716], [221.55283, 221.48502, 213.44911], [221.12857, 221.24738, 213.23395], [220.76405, 220.92285, 212.88655], [220.37428, 220.55666, 212.36052], [220.02591, 220.14854, 211.68695], [219.67354, 219.66013, 210.86441], [219.31143, 219.10959, 209.93048], [219.05827, 218.59978, 208.71603], [218.86418, 218.17699, 204.88757], [218.67836, 217.58351, 201.41728], [218.61922, 217.45003, 199.41467], [218.81314, 217.10815, 200.6058 ], [219.00188, 216.90114, 202.06958], [219.58716, 217.05151, 203.64798], [220.07086, 217.46791, 205.44212], [220.14432, 217.79782, 206.75606], [220.10417, 217.91089, 207.8027 ], [220.27509, 218.02267, 208.39905], [220.46239, 218.16553, 209.34784], [220.71407, 218.57372, 210.57094]], dtype=float32) - FREQI(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- fraction
- long_name :
- Fractional occurrence of ice
- basename :
- FREQI
array([[[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., ... ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - FREQL(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- fraction
- long_name :
- Fractional occurrence of liquid
- basename :
- FREQL
array([[[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., ... ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - FREQR(time, lev, ncol)float320.0001 0.0001 0.0005159 ... 0.0 0.0
- mdims :
- 9
- units :
- fraction
- long_name :
- Fractional occurrence of rain
- basename :
- FREQR
array([[[9.9999997e-05, 9.9999997e-05, 5.1592407e-04], [9.9999997e-05, 9.9999997e-05, 3.8299891e-03], [1.4467775e-04, 5.2718641e-03, 4.5738880e-02], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[9.9999997e-05, 9.9999997e-05, 6.3581247e-04], [9.9999997e-05, 9.9999997e-05, 5.1842085e-03], [1.4255832e-04, 1.5475810e-02, 6.3984610e-02], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[9.9999997e-05, 9.9999997e-05, 1.1370420e-03], [9.9999997e-05, 2.3446756e-03, 9.8446077e-03], [9.9999997e-05, 3.0964246e-02, 1.1348072e-01], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[9.9999997e-05, 3.0046186e-04, 0.0000000e+00], [1.2313537e-04, 6.4018444e-04, 0.0000000e+00], [4.1422047e-04, 2.1369918e-03, 9.9999997e-05], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[9.9999997e-05, 1.1765216e-04, 9.9999997e-05], [1.2218198e-04, 2.5570864e-04, 9.9999997e-05], [4.1664098e-04, 8.7710691e-04, 9.9999997e-05], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - FREQS(time, lev, ncol)float320.0 0.0001 0.0005159 ... 0.0 0.0
- mdims :
- 9
- units :
- fraction
- long_name :
- Fractional occurrence of snow
- basename :
- FREQS
array([[[0.0000000e+00, 9.9999997e-05, 5.1592407e-04], [0.0000000e+00, 9.9999997e-05, 3.8299891e-03], [0.0000000e+00, 5.2718641e-03, 4.5738880e-02], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 9.9999997e-05, 6.3581247e-04], [0.0000000e+00, 9.9999997e-05, 5.1842085e-03], [0.0000000e+00, 1.5475810e-02, 6.3984610e-02], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 9.9999997e-05, 1.1370420e-03], [0.0000000e+00, 2.3446756e-03, 9.8446077e-03], [0.0000000e+00, 3.0964246e-02, 1.1348072e-01], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[9.9999997e-05, 3.0046186e-04, 9.9999997e-05], [1.2313537e-04, 6.4018444e-04, 9.9999997e-05], [4.1422047e-04, 2.1369918e-03, 9.9999997e-05], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[9.9999997e-05, 1.1765216e-04, 9.9999997e-05], [1.2218198e-04, 2.5570864e-04, 9.9999997e-05], [4.1664098e-04, 8.7710691e-04, 9.9999997e-05], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - FSDS(time, ncol)float320.2678 1.545 5.795 ... 7.52 16.71
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Downwelling solar flux at surface
- basename :
- FSDS
array([[2.6784062e-01, 1.5451398e+00, 5.7951522e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 1.6117154e-03], [4.0632766e-01, 7.9035702e+00, 8.9394522e+00], [3.1724916e+00, 1.4155913e+01, 2.5688362e+01], [5.0563054e+00, 1.6244247e+01, 3.2292610e+01], [4.6395631e+00, 1.2578314e+01, 2.8419497e+01], [2.1743336e+00, 7.5203700e+00, 1.6713949e+01]], dtype=float32) - FSDSC(time, ncol)float320.7079 4.409 10.67 ... 24.9 37.11
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Clearsky downwelling solar flux at surface
- basename :
- FSDSC
array([[7.0791477e-01, 4.4091682e+00, 1.0670688e+01], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 1.8038332e-03], [5.0365204e-01, 9.1871767e+00, 1.3504834e+01], [1.0995711e+01, 3.3976185e+01, 4.4699913e+01], [2.2566004e+01, 4.8407459e+01, 6.1969952e+01], [2.1745045e+01, 4.4037598e+01, 5.8645184e+01], [9.6550722e+00, 2.4901352e+01, 3.7107403e+01]], dtype=float32) - FSNS(time, ncol)float320.2518 1.13 1.735 ... 5.332 4.504
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Net solar flux at surface
- basename :
- FSNS
array([[ 0.2517702, 1.1300002, 1.7348864], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 2.7061872], [ 2.9821422, 10.223478 , 7.5255494], [ 4.7529273, 11.6351385, 9.145517 ], [ 4.361189 , 8.948196 , 7.862798 ], [ 2.0438735, 5.3315816, 4.50407 ]], dtype=float32) - FSNT(time, ncol)float321.5 8.036 14.08 ... 24.17 27.9
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Net solar flux at top of model
- basename :
- FSNT
array([[1.4996370e+00, 8.0357513e+00, 1.4078048e+01], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 7.5198774e-04], [8.5382235e-01, 9.8820763e+00, 1.4920725e+01], [1.4783431e+01, 3.2216465e+01, 3.2896168e+01], [2.2289629e+01, 3.9315361e+01, 4.0602570e+01], [2.2081701e+01, 3.4989517e+01, 3.8112064e+01], [1.3228238e+01, 2.4170631e+01, 2.7896496e+01]], dtype=float32) - FSNTOAC(time, ncol)float321.893 10.0 16.48 ... 38.74 40.46
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Clearsky net solar flux at top of atmosphere
- basename :
- FSNTOAC
array([[1.8931845e+00, 1.0001142e+01, 1.6477505e+01], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 1.5820465e-03], [8.5347366e-01, 1.1624482e+01, 1.9533609e+01], [2.3529543e+01, 4.8126282e+01, 4.6036076e+01], [4.0451889e+01, 6.3026707e+01, 5.8386879e+01], [3.9554085e+01, 5.9255810e+01, 5.6349354e+01], [2.1338987e+01, 3.8740356e+01, 4.0459286e+01]], dtype=float32) - FSUTOA(time, ncol)float321.283 7.505 16.68 ... 39.81 55.82
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Upwelling solar flux at top of atmosphere
- basename :
- FSUTOA
array([[1.2828287e+00, 7.5045028e+00, 1.6678265e+01], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 3.7065360e-03], [1.0233163e+00, 1.7630880e+01, 2.2143217e+01], [2.0395973e+01, 4.6636490e+01, 5.9763546e+01], [3.8389824e+01, 6.3681736e+01, 7.9275940e+01], [3.7563786e+01, 6.2940922e+01, 7.8719780e+01], [1.8902985e+01, 3.9808807e+01, 5.5816326e+01]], dtype=float32) - GCLDLWP(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/m2
- long_name :
- Grid-box cloud water path
- basename :
- GCLDLWP
array([[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 7.0755270e-08], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 4.6564488e-12, 1.8506066e-07], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 5.2647921e-12, 6.1114486e-07], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - ICE_ICLD_VISTAU(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- Sampling_Sequence :
- rad_lwsw
- units :
- 1
- long_name :
- Ice in-cloud extinction visible sw optical depth
- basename :
- ICE_ICLD_VISTAU
array([[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 3.0424377e-05], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan]], [[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - ICIMR(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Prognostic in-cloud ice mixing ratio
- basename :
- ICIMR
array([[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 1.7145567e-08], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 3.3080611e-12, 3.2037882e-08], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 1.8682590e-12, 5.9616909e-08], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - ICIMRST(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Prognostic in-stratus ice mixing ratio
- basename :
- ICIMRST
array([[[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., ... ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - ICINC(time, lev, ncol)float321.442 0.4038 0.08901 ... 0.0 0.0
- mdims :
- 9
- units :
- m-3
- long_name :
- Prognostic in-cloud ice number conc
- basename :
- ICINC
array([[[1.44229817e+00, 4.03844982e-01, 8.90069306e-02], [5.44832182e+00, 5.45721710e-01, 8.02163720e-01], [7.74697351e+00, 3.57159786e-02, 4.23926735e+01], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[1.00963342e+00, 3.53216529e-01, 1.31513819e-01], [4.02839184e+00, 4.74126965e-01, 1.20813167e+00], [5.08871460e+00, 1.14117423e-02, 6.49467773e+01], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[1.99138200e+00, 1.59541696e-01, 1.90097392e-01], [2.08007121e+00, 1.99129973e-02, 1.78067136e+00], [2.81348348e+00, 7.83493444e-02, 9.92995148e+01], ..., ... ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[3.98051634e-04, 1.92199589e-03, 3.32501456e-02], [7.64984346e-04, 1.92228542e-03, 3.76256518e-02], [7.58464355e-03, 7.07935616e-02, 2.60702036e-02], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[3.21464118e-04, 8.61531496e-03, 9.67848953e-03], [6.65316300e-04, 1.19816974e-01, 1.75180081e-02], [5.06022293e-03, 1.38874984e+00, 4.34276760e-02], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]]], dtype=float32) - ICLDIWP(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/m2
- long_name :
- In-cloud ice water path
- basename :
- ICLDIWP
array([[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 1.5469393e-06], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 3.0088565e-10, 2.8922682e-06], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 1.7002810e-10, 5.3854510e-06], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - ICLDTWP(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/m2
- long_name :
- In-cloud cloud total water path (liquid and ice)
- basename :
- ICLDTWP
array([[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 1.5469393e-06], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 3.0088565e-10, 2.8922682e-06], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 1.7002810e-10, 5.3854510e-06], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - ICWMRST(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Prognostic in-stratus water mixing ratio
- basename :
- ICWMRST
array([[[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., ... ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - ICWNC(time, lev, ncol)float321.347e-08 1.335e-08 ... 0.0 0.0
- mdims :
- 9
- units :
- m-3
- long_name :
- Prognostic in-cloud water number conc
- basename :
- ICWNC
array([[[1.34728992e-08, 1.33524880e-08, 2.58322053e-09], [1.34277940e-08, 1.33074174e-08, 3.46688594e-10], [9.23126020e-09, 2.51064614e-10, 2.88648341e-11], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[1.34724161e-08, 1.33527260e-08, 2.09655404e-09], [1.34271447e-08, 1.33077291e-08, 2.56170224e-10], [9.36786382e-09, 8.55291393e-11, 2.06367961e-11], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[1.34719711e-08, 1.33541089e-08, 1.17259857e-09], [1.34265061e-08, 5.67640657e-10, 1.34928277e-10], [1.33537785e-08, 4.27521386e-11, 1.16382121e-11], ..., ... ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[1.30224080e-08, 4.39319070e-09, 1.32836417e-08], [1.05397548e-08, 2.05498063e-09, 1.32369387e-08], [2.15117186e-02, 6.58351853e-02, 1.31608235e-08], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[1.30222677e-08, 1.11831246e-08, 1.32281697e-08], [1.06219353e-08, 5.12813303e-09, 1.31819435e-08], [6.54823482e-02, 6.18341625e-01, 2.29554325e-02], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]]], dtype=float32) - IWC(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/m3
- long_name :
- Grid box average ice water content
- basename :
- IWC
array([[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 1.0353614e-09], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 6.7763273e-14, 2.7068061e-09], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 7.6580113e-14, 8.9351095e-09], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - LIQ_ICLD_VISTAU(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- Sampling_Sequence :
- rad_lwsw
- units :
- 1
- long_name :
- Liquid in-cloud extinction visible sw optical depth
- basename :
- LIQ_ICLD_VISTAU
array([[[ 0., 0., 0.], [ 0., 0., 0.], [ 0., 0., 0.], ..., [ 0., 0., 0.], [ 0., 0., 0.], [ 0., 0., 0.]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., ... ..., [ 0., 0., 0.], [ 0., 0., 0.], [ 0., 0., 0.]], [[ 0., 0., 0.], [ 0., 0., 0.], [ 0., 0., 0.], ..., [ 0., 0., 0.], [ 0., 0., 0.], [ 0., 0., 0.]], [[ 0., 0., 0.], [ 0., 0., 0.], [ 0., 0., 0.], ..., [ 0., 0., 0.], [ 0., 0., 0.], [ 0., 0., 0.]]], dtype=float32) - LWC(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/m3
- long_name :
- Grid box average liquid water content
- basename :
- LWC
array([[[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., ... ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - LWCF(time, ncol)float3221.72 26.09 21.48 ... 61.24 52.31
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Longwave cloud forcing
- basename :
- LWCF
array([[21.720066, 26.088118, 21.480127], [21.628384, 25.18713 , 20.91648 ], [22.117128, 24.73137 , 20.36388 ], [22.89166 , 24.308146, 19.924183], [23.587181, 23.923824, 19.504282], [28.35875 , 23.812637, 18.908743], [35.497795, 23.961866, 18.399971], [40.291237, 24.312006, 18.063877], [43.64851 , 24.454327, 17.741922], [45.169262, 25.282734, 17.368832], [50.111767, 27.606243, 17.074776], [54.338596, 31.096115, 14.200049], [55.69818 , 33.043224, 14.64996 ], [57.89926 , 36.51449 , 16.067335], [58.376057, 37.259144, 18.139975], [67.68696 , 35.88557 , 19.05946 ], [64.31541 , 54.427338, 31.430033], [66.07005 , 60.166695, 46.220898], [59.75964 , 62.18478 , 46.8051 ], [54.95062 , 58.25685 , 43.912178], [58.839977, 55.005943, 43.295322], [64.75803 , 51.796204, 42.121468], [61.02838 , 59.613667, 48.58198 ], [53.625328, 61.23687 , 52.313988]], dtype=float32) - NSNOW(time, lev, ncol)float320.009382 41.2 235.6 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- m-3
- long_name :
- Diagnostic grid-mean snow number conc
- basename :
- NSNOW
array([[[9.38184839e-03, 4.11979256e+01, 2.35579956e+02], [9.57082864e-03, 5.18728752e+01, 3.83739838e+02], [1.00646671e-02, 6.08924599e+01, 4.66344238e+02], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[6.50361134e-03, 4.35848312e+01, 2.23704865e+02], [6.65427418e-03, 5.53294296e+01, 3.72962708e+02], [6.99624699e-03, 6.36766548e+01, 4.41380005e+02], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[4.33164136e-03, 4.54658966e+01, 2.36037552e+02], [4.50564316e-03, 4.72252884e+01, 4.04535339e+02], [4.73473966e-03, 5.43587074e+01, 4.55066010e+02], ..., ... ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[2.39686005e+02, 2.79277405e+02, 1.11828718e+01], [2.66172485e+02, 3.02869415e+02, 1.56308565e+01], [3.26526886e+02, 3.59139313e+02, 2.75170879e+01], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[2.04049454e+02, 4.76226440e+02, 1.63210236e+02], [2.23062576e+02, 5.14286621e+02, 1.89062881e+02], [2.66488892e+02, 6.05180115e+02, 2.47630005e+02], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]]], dtype=float32) - NUMICE(time, lev, ncol)float321.443e-10 4.04e-11 ... 1.627e-22
- mdims :
- 9
- units :
- 1/kg
- long_name :
- Grid box averaged cloud ice number
- basename :
- NUMICE
array([[[1.4427604e-10, 4.0397442e-11, 4.5935533e-11], [5.4500676e-10, 5.4589663e-11, 3.0732625e-09], [1.1211739e-09, 1.8835014e-10, 1.9396148e-06], ..., [3.6240230e-22, 3.6060316e-22, 3.5928719e-22], [2.3979228e-22, 2.3632162e-22, 2.3534816e-22], [1.6293956e-22, 1.6029121e-22, 1.5945670e-22]], [[1.0099570e-10, 3.5332969e-11, 8.3644924e-11], [4.0296827e-10, 4.7427888e-11, 6.2652141e-09], [7.2567113e-10, 1.7666255e-10, 4.1569256e-06], ..., [3.6164436e-22, 3.6098835e-22, 3.5984714e-22], [2.3812298e-22, 2.3453317e-22, 2.3367552e-22], [1.6258192e-22, 1.5938100e-22, 1.5855015e-22]], [[1.9920202e-10, 1.5959281e-11, 2.1621795e-10], [2.0807377e-10, 4.6704484e-11, 1.7535628e-08], [2.8143854e-10, 2.4268056e-09, 1.1272190e-05], ..., ... ..., [3.7164706e-22, 3.6889431e-22, 3.6758040e-22], [2.4962020e-22, 2.4857679e-22, 2.4783059e-22], [1.6577590e-22, 1.6392872e-22, 1.6340139e-22]], [[3.9817918e-14, 5.7767154e-13, 3.3260801e-12], [9.4226805e-14, 1.2310117e-12, 3.7637710e-12], [3.1427211e-12, 1.5133375e-10, 2.6078556e-12], ..., [3.7138014e-22, 3.6890665e-22, 3.6765932e-22], [2.4843858e-22, 2.4683294e-22, 2.4619417e-22], [1.6556638e-22, 1.6368841e-22, 1.6339610e-22]], [[3.2156711e-14, 1.0139351e-12, 9.6815904e-13], [8.1315719e-14, 3.0648050e-11, 1.7523620e-12], [2.1089719e-12, 1.2184724e-09, 4.3441591e-12], ..., [3.7005695e-22, 3.6743010e-22, 3.6622339e-22], [2.4670197e-22, 2.4452449e-22, 2.4382497e-22], [1.6555971e-22, 1.6333287e-22, 1.6272291e-22]]], dtype=float32) - NUMIMM10sBC(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- #/m3
- long_name :
- Ice Number Concentration due to imm freezing by bc in Mixed Clouds during 10-s period
- basename :
- NUMIMM10sBC
array([[[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., ... ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - NUMIMM10sDST(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- #/m3
- long_name :
- Ice Number Concentration due to imm freezing by dst in Mixed Clouds during 10-s period
- basename :
- NUMIMM10sDST
array([[[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., ... ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - NUMLIQ(time, lev, ncol)float321.348e-18 1.336e-18 ... 1.627e-22
- mdims :
- 9
- units :
- 1/kg
- long_name :
- Grid box averaged cloud liquid number
- basename :
- NUMLIQ
array([[[1.3477215e-18, 1.3356769e-18, 1.3331727e-18], [1.3432097e-18, 1.3311682e-18, 1.3282390e-18], [1.3359860e-18, 1.3240028e-18, 1.3206682e-18], ..., [3.6240230e-22, 3.6060316e-22, 3.5928719e-22], [2.3979228e-22, 2.3632162e-22, 2.3534816e-22], [1.6293956e-22, 1.6029121e-22, 1.5945670e-22]], [[1.3476734e-18, 1.3357004e-18, 1.3334423e-18], [1.3431448e-18, 1.3311993e-18, 1.3284655e-18], [1.3358949e-18, 1.3240568e-18, 1.3208605e-18], ..., [3.6164436e-22, 3.6098835e-22, 3.5984714e-22], [2.3812298e-22, 2.3453317e-22, 2.3367552e-22], [1.6258192e-22, 1.5938100e-22, 1.5855015e-22]], [[1.3476288e-18, 1.3358388e-18, 1.3337209e-18], [1.3430808e-18, 1.3313598e-18, 1.3287415e-18], [1.3358058e-18, 1.3242119e-18, 1.3211359e-18], ..., ... ..., [3.7164706e-22, 3.6889431e-22, 3.6758040e-22], [2.4962020e-22, 2.4857679e-22, 2.4783059e-22], [1.6577590e-22, 1.6392872e-22, 1.6340139e-22]], [[1.3026580e-18, 1.3204093e-18, 1.3287898e-18], [1.2982324e-18, 1.3159882e-18, 1.3241181e-18], [8.9134490e-12, 1.4073435e-10, 1.3165041e-18], ..., [3.7138014e-22, 3.6890665e-22, 3.6765932e-22], [2.4843858e-22, 2.4683294e-22, 2.4619417e-22], [1.6556638e-22, 1.6368841e-22, 1.6339610e-22]], [[1.3026440e-18, 1.3161404e-18, 1.3232408e-18], [1.2982249e-18, 1.3117280e-18, 1.3186167e-18], [2.7291374e-11, 5.4252552e-10, 2.2962786e-12], ..., [3.7005695e-22, 3.6743010e-22, 3.6622339e-22], [2.4670197e-22, 2.4452449e-22, 2.4382497e-22], [1.6555971e-22, 1.6333287e-22, 1.6272291e-22]]], dtype=float32) - NUMRAI(time, lev, ncol)float320.001946 0.003069 ... 1.627e-22
- mdims :
- 9
- units :
- 1/kg
- long_name :
- Grid box averaged rain number
- basename :
- NUMRAI
array([[[1.94640318e-03, 3.06928414e-03, 4.85477922e-03], [4.85288352e-03, 5.32037253e-03, 7.77697936e-03], [1.68888103e-02, 5.87743800e-03, 1.83627941e-02], ..., [3.62402296e-22, 3.60603163e-22, 3.59287192e-22], [2.39792283e-22, 2.36321624e-22, 2.35348157e-22], [1.62939561e-22, 1.60291210e-22, 1.59456696e-22]], [[1.59615313e-03, 2.29752297e-03, 4.71569365e-03], [4.03811364e-03, 8.10895581e-03, 7.20570190e-03], [1.43019333e-02, 1.93274673e-02, 1.71663277e-02], ..., [3.61644358e-22, 3.60988354e-22, 3.59847144e-22], [2.38122978e-22, 2.34533169e-22, 2.33675519e-22], [1.62581923e-22, 1.59381003e-22, 1.58550149e-22]], [[2.02719573e-04, 5.97174838e-03, 4.92139859e-03], [1.21054635e-03, 9.40920506e-03, 9.26272012e-03], [4.09822864e-03, 2.40395684e-02, 1.54526606e-02], ..., ... ..., [3.71647064e-22, 3.68894306e-22, 3.67580404e-22], [2.49620203e-22, 2.48576786e-22, 2.47830587e-22], [1.65775901e-22, 1.63928717e-22, 1.63401391e-22]], [[2.60633649e-03, 1.19531888e-03, 4.49929712e-06], [5.13654109e-03, 2.22072774e-03, 1.00217485e-05], [1.43029094e-02, 5.65801375e-03, 3.60330196e-05], ..., [3.71380139e-22, 3.68906650e-22, 3.67659316e-22], [2.48438578e-22, 2.46832936e-22, 2.46194174e-22], [1.65566379e-22, 1.63688411e-22, 1.63396103e-22]], [[3.26708914e-03, 2.11402751e-03, 4.37628973e-04], [6.51298091e-03, 3.79784522e-03, 8.62879097e-04], [1.85913779e-02, 9.22998507e-03, 2.51152948e-03], ..., [3.70056948e-22, 3.67430104e-22, 3.66223387e-22], [2.46701973e-22, 2.44524490e-22, 2.43824966e-22], [1.65559715e-22, 1.63332868e-22, 1.62722908e-22]]], dtype=float32) - NUMSNO(time, lev, ncol)float321.137e-08 3.324e-05 ... 1.627e-22
- mdims :
- 9
- units :
- 1/kg
- long_name :
- Grid box averaged snow number
- basename :
- NUMSNO
array([[[1.1368726e-08, 3.3238161e-05, 1.8056590e-04], [1.1456228e-08, 3.4081244e-05, 4.0867305e-04], [1.1541645e-08, 7.0514223e-05, 4.7684051e-04], ..., [3.6240230e-22, 3.6060316e-22, 3.5928719e-22], [2.3979228e-22, 2.3632162e-22, 2.3534816e-22], [1.6293956e-22, 1.6029121e-22, 1.5945670e-22]], [[7.9196596e-09, 3.4384142e-05, 1.7710467e-04], [8.0496401e-09, 3.5155077e-05, 4.0344929e-04], [8.2013223e-09, 7.6933189e-05, 4.5493318e-04], ..., [3.6164436e-22, 3.6098835e-22, 3.5984714e-22], [2.3812298e-22, 2.3453317e-22, 2.3367552e-22], [1.6258192e-22, 1.5938100e-22, 1.5855015e-22]], [[4.7537139e-09, 2.6097894e-05, 1.8486373e-04], [5.1927502e-09, 5.4521974e-05, 4.3413608e-04], [5.4241114e-09, 6.0083279e-05, 4.6168186e-04], ..., ... ..., [3.7164706e-22, 3.6889431e-22, 3.6758040e-22], [2.4962020e-22, 2.4857679e-22, 2.4783059e-22], [1.6577590e-22, 1.6392872e-22, 1.6340139e-22]], [[1.7743109e-04, 2.5383639e-04, 6.2836448e-06], [2.1220440e-04, 2.9334472e-04, 1.0544682e-05], [2.8342707e-04, 3.6953844e-04, 2.2833710e-05], ..., [3.7138014e-22, 3.6890665e-22, 3.6765932e-22], [2.4843858e-22, 2.4683294e-22, 2.4619417e-22], [1.6556638e-22, 1.6368841e-22, 1.6339610e-22]], [[1.5777651e-04, 4.0387263e-04, 1.5086352e-04], [1.8405647e-04, 4.6440834e-04, 1.9219190e-04], [2.3554526e-04, 5.7899143e-04, 2.8078773e-04], ..., [3.7005695e-22, 3.6743010e-22, 3.6622339e-22], [2.4670197e-22, 2.4452449e-22, 2.4382497e-22], [1.6555971e-22, 1.6333287e-22, 1.6272291e-22]]], dtype=float32) - OMEGA(time, lev, ncol)float320.01072 -0.0316 ... 5.859e-05
- mdims :
- 9
- units :
- Pa/s
- long_name :
- Vertical velocity (pressure)
- basename :
- OMEGA
array([[[ 1.07247764e-02, -3.16038355e-02, -3.81121896e-02], [ 1.64787862e-02, -3.49201821e-02, -5.44950329e-02], [ 2.51672845e-02, -3.44999917e-02, -5.95921129e-02], ..., [ 1.54458408e-04, 6.23800452e-06, -5.19820715e-05], [ 2.00939015e-04, 3.66939421e-05, -2.99668409e-06], [ 9.20130624e-05, 4.28405983e-05, 3.21103835e-05]], [[ 6.95641851e-03, -3.49932127e-02, -3.81200500e-02], [ 1.31674763e-02, -3.79534923e-02, -5.49081862e-02], [ 2.25521848e-02, -3.68079618e-02, -5.92842363e-02], ..., [ 1.15136179e-04, -3.81073801e-06, -3.96243740e-05], [ 1.23753503e-04, -2.36360829e-05, -3.57929166e-05], [ 5.22241717e-05, -1.41900989e-06, -2.32743992e-06]], [[ 1.33657139e-02, -2.31908560e-02, -2.24495884e-02], [ 1.99950822e-02, -2.55774856e-02, -3.89924012e-02], [ 3.00504137e-02, -2.34957300e-02, -4.25684154e-02], ..., ... [ 8.11124846e-05, 1.00271471e-04, 1.50428139e-04], [ 7.08494554e-05, 1.83773664e-05, 4.49113759e-05], [ 2.72248344e-05, -1.25814422e-05, -8.57438772e-06]], [[-4.71803844e-02, -2.61060540e-02, 6.21933192e-02], [-6.34271577e-02, -3.15920375e-02, 6.34866282e-02], [-8.87005329e-02, -4.59739566e-02, 5.00574708e-02], ..., [ 1.59781630e-04, 1.44040299e-04, 1.77030641e-04], [ 1.36242481e-04, 1.09185712e-04, 1.15848736e-04], [ 4.82851574e-05, 3.62434221e-05, 2.73728801e-05]], [[-3.41812223e-02, -2.60860939e-02, 5.49512506e-02], [-4.87268195e-02, -3.64790149e-02, 4.96051274e-02], [-7.09889680e-02, -5.72536364e-02, 3.00648753e-02], ..., [ 1.09345077e-04, 1.29590844e-04, 1.45226717e-04], [ 1.07589483e-04, 1.49450760e-04, 1.52054767e-04], [ 3.50033952e-05, 6.28787602e-05, 5.85891103e-05]]], dtype=float32) - PRAO(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg/s
- long_name :
- Accretion of cloud water by rain
- basename :
- PRAO
array([[[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., ... ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - PRCO(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg/s
- long_name :
- Autoconversion of cloud water
- basename :
- PRCO
array([[[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., ... ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - PRECC(time, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- units :
- m/s
- long_name :
- Convective precipitation rate (liq + ice)
- basename :
- PRECC
array([[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], dtype=float32) - PRECL(time, ncol)float325.93e-09 1.951e-09 ... 8.1e-08
- units :
- m/s
- long_name :
- Large-scale (stable) precipitation rate (liq + ice)
- basename :
- PRECL
array([[5.9299561e-09, 1.9512592e-09, 2.9270997e-09], [4.4443103e-09, 1.9518263e-09, 3.1760576e-09], [9.7244657e-10, 1.6456754e-09, 3.2875216e-09], [8.1694318e-10, 7.0188005e-10, 1.8406706e-09], [9.0446062e-10, 1.6339069e-09, 3.3472345e-09], [1.0929112e-09, 1.0531480e-09, 3.1833314e-09], [2.4958691e-09, 6.6675793e-10, 2.8726390e-09], [4.1774659e-09, 6.2743033e-10, 1.8212674e-09], [7.1855770e-09, 8.9150648e-10, 1.6596237e-09], [1.2024576e-08, 1.4296134e-09, 2.0578177e-09], [1.6044538e-08, 1.2633652e-09, 1.8560489e-09], [2.1517959e-08, 1.1870004e-09, 1.9995150e-09], [3.3073018e-08, 3.1303118e-09, 1.1309258e-09], [4.7411962e-08, 5.0197793e-09, 2.4829050e-09], [6.0607832e-08, 8.8771026e-09, 5.6539911e-09], [7.2457844e-08, 1.2414785e-08, 8.4116776e-09], [8.8227878e-08, 1.7837364e-08, 1.2592615e-08], [1.2422709e-07, 2.6795512e-08, 1.9443855e-08], [1.4315874e-07, 3.7370210e-08, 2.6760240e-08], [1.5355623e-07, 5.8406521e-08, 3.9917239e-08], [1.4134434e-07, 6.9522059e-08, 4.9989648e-08], [1.2525558e-07, 7.6711558e-08, 5.8604616e-08], [1.0492104e-07, 9.0306266e-08, 7.0798009e-08], [7.9961993e-08, 1.2207617e-07, 8.1002284e-08]], dtype=float32) - PRECT(time, ncol)float325.93e-09 1.951e-09 ... 8.1e-08
- units :
- m/s
- long_name :
- Total (convective and large-scale) precipitation rate (liq + ice)
- basename :
- PRECT
array([[5.9299561e-09, 1.9512592e-09, 2.9270997e-09], [4.4443103e-09, 1.9518263e-09, 3.1760576e-09], [9.7244657e-10, 1.6456754e-09, 3.2875216e-09], [8.1694318e-10, 7.0188005e-10, 1.8406706e-09], [9.0446062e-10, 1.6339069e-09, 3.3472345e-09], [1.0929112e-09, 1.0531480e-09, 3.1833314e-09], [2.4958691e-09, 6.6675793e-10, 2.8726390e-09], [4.1774659e-09, 6.2743033e-10, 1.8212674e-09], [7.1855770e-09, 8.9150648e-10, 1.6596237e-09], [1.2024576e-08, 1.4296134e-09, 2.0578177e-09], [1.6044538e-08, 1.2633652e-09, 1.8560489e-09], [2.1517959e-08, 1.1870004e-09, 1.9995150e-09], [3.3073018e-08, 3.1303118e-09, 1.1309258e-09], [4.7411962e-08, 5.0197793e-09, 2.4829050e-09], [6.0607832e-08, 8.8771026e-09, 5.6539911e-09], [7.2457844e-08, 1.2414785e-08, 8.4116776e-09], [8.8227878e-08, 1.7837364e-08, 1.2592615e-08], [1.2422709e-07, 2.6795512e-08, 1.9443855e-08], [1.4315874e-07, 3.7370210e-08, 2.6760240e-08], [1.5355623e-07, 5.8406521e-08, 3.9917239e-08], [1.4134434e-07, 6.9522059e-08, 4.9989648e-08], [1.2525558e-07, 7.6711558e-08, 5.8604616e-08], [1.0492104e-07, 9.0306266e-08, 7.0798009e-08], [7.9961993e-08, 1.2207617e-07, 8.1002284e-08]], dtype=float32) - PS(time, ncol)float321.034e+05 1.03e+05 ... 1.018e+05
- units :
- Pa
- long_name :
- Surface pressure
- basename :
- PS
array([[103446.984, 103024.234, 102308.47 ], [103478.01 , 103074.67 , 102363.61 ], [103508.875, 103130.77 , 102424.13 ], [103528.22 , 103179.96 , 102479.14 ], [103528.875, 103212.375, 102514.34 ], [103511.91 , 103237.81 , 102545.625], [103491.79 , 103260.92 , 102571.195], [103459.57 , 103280.01 , 102593.38 ], [103415.914, 103296.08 , 102613.805], [103372.62 , 103320.23 , 102644.82 ], [103303.18 , 103323.2 , 102655.914], [103214.25 , 103312.4 , 102656.41 ], [103113.4 , 103288.53 , 102648.57 ], [102988.73 , 103240.195, 102616.97 ], [102853.82 , 103187.5 , 102580.79 ], [102712.39 , 103118.41 , 102529.76 ], [102578.14 , 103054.86 , 102484.74 ], [102436.42 , 102969.05 , 102419.984], [102305.47 , 102868.5 , 102342.445], [102190.47 , 102768.414, 102259.03 ], [102102.234, 102665.336, 102175.945], [102008.81 , 102531.46 , 102064.64 ], [101924.33 , 102378.85 , 101931.945], [101875.07 , 102241.84 , 101811.42 ]], dtype=float32) - PSL(time, ncol)float321.034e+05 1.033e+05 ... 1.028e+05
- units :
- Pa
- long_name :
- Sea level pressure
- basename :
- PSL
array([[103447.58 , 103290.86 , 103336.76 ], [103478.61 , 103341.305, 103392.1 ], [103509.48 , 103397.43 , 103452.84 ], [103528.82 , 103446.63 , 103508.08 ], [103529.48 , 103479.04 , 103543.36 ], [103512.5 , 103504.46 , 103574.766], [103492.39 , 103527.57 , 103600.555], [103460.164, 103546.64 , 103622.97 ], [103416.51 , 103562.7 , 103643.79 ], [103373.21 , 103586.91 , 103675.45 ], [103303.77 , 103589.875, 103687.31 ], [103214.84 , 103579.02 , 103690.61 ], [103113.984, 103555.08 , 103686.664], [102989.31 , 103506.64 , 103658.15 ], [102854.41 , 103453.875, 103622.88 ], [102712.98 , 103384.695, 103571.61 ], [102578.73 , 103321.05 , 103525.44 ], [102437. , 103235.07 , 103458.51 ], [102306.055, 103134.266, 103378.945], [102191.05 , 103033.84 , 103293.39 ], [102102.81 , 102930.336, 103208.016], [102009.39 , 102795.836, 103093.47 ], [101924.91 , 102642.43 , 102956.85 ], [101875.65 , 102504.57 , 102832.04 ]], dtype=float32) - Q(time, lev, ncol)float320.001912 0.001889 ... 2.172e-06
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Specific humidity
- basename :
- Q
array([[[1.9123822e-03, 1.8890033e-03, 1.8980128e-03], [1.9053145e-03, 1.8854522e-03, 1.8866546e-03], [1.8975340e-03, 1.8749102e-03, 1.8772145e-03], ..., [2.1726269e-06, 2.1736416e-06, 2.1712017e-06], [2.1540654e-06, 2.1414792e-06, 2.1438318e-06], [2.1281078e-06, 2.1633541e-06, 2.1900128e-06]], [[1.9103336e-03, 1.9377643e-03, 1.9286064e-03], [1.9034640e-03, 1.9344364e-03, 1.9189988e-03], [1.8959978e-03, 1.9258412e-03, 1.9119459e-03], ..., [2.1690369e-06, 2.1703786e-06, 2.1680453e-06], [2.1528379e-06, 2.1511430e-06, 2.1582223e-06], [2.1399544e-06, 2.1747974e-06, 2.2052213e-06]], [[1.9023425e-03, 1.9875264e-03, 1.9635749e-03], [1.8965562e-03, 1.9854268e-03, 1.9558358e-03], [1.8902243e-03, 1.9770563e-03, 1.9512721e-03], ..., ... ..., [2.1630269e-06, 2.1623748e-06, 2.1620565e-06], [2.1642138e-06, 2.1635331e-06, 2.1655483e-06], [2.1451160e-06, 2.1730709e-06, 2.1797139e-06]], [[2.9548192e-03, 2.5196110e-03, 2.0498680e-03], [2.9498693e-03, 2.5149763e-03, 2.0459704e-03], [2.9350056e-03, 2.4962116e-03, 2.0416782e-03], ..., [2.1639592e-06, 2.1618685e-06, 2.1608093e-06], [2.1660073e-06, 2.1666090e-06, 2.1642998e-06], [2.1596043e-06, 2.1666706e-06, 2.1773180e-06]], [[2.9335942e-03, 2.6397710e-03, 2.2063514e-03], [2.9285832e-03, 2.6347223e-03, 2.2037136e-03], [2.9139600e-03, 2.6166078e-03, 2.2012063e-03], ..., [2.1643350e-06, 2.1626647e-06, 2.1627166e-06], [2.1666149e-06, 2.1697833e-06, 2.1664714e-06], [2.1706082e-06, 2.1635290e-06, 2.1724954e-06]]], dtype=float32) - QRAIN(time, lev, ncol)float321.48e-06 2.503e-07 ... 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Diagnostic grid-mean rain mixing ratio
- basename :
- QRAIN
array([[[1.47966000e-06, 2.50254345e-07, 2.84792861e-07], [1.91441200e-06, 4.17997796e-07, 5.09184190e-07], [3.90325522e-06, 1.05612241e-06, 1.30043668e-06], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[1.14005411e-06, 2.26072999e-07, 3.58391020e-07], [1.47378182e-06, 4.62719555e-07, 6.06988294e-07], [3.04445302e-06, 1.22901929e-06, 1.55147291e-06], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[1.90476987e-06, 8.48022637e-07, 3.41689798e-07], [2.47010189e-06, 1.58484420e-06, 7.21994581e-07], [3.57789440e-06, 3.58649004e-06, 1.74281968e-06], ..., ... ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[9.37349614e-06, 3.11662552e-06, 8.23223729e-08], [9.98301130e-06, 3.33647631e-06, 9.71327694e-08], [1.07663554e-05, 3.79331527e-06, 1.35249763e-07], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[9.32748571e-06, 6.27552890e-06, 1.77809034e-06], [1.00918205e-05, 6.68127404e-06, 1.98974976e-06], [1.14939548e-05, 7.51141260e-06, 2.49171876e-06], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]]], dtype=float32) - QSNOW(time, lev, ncol)float322.142e-09 1.252e-06 ... 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Diagnostic grid-mean snow mixing ratio
- basename :
- QSNOW
array([[[2.1419162e-09, 1.2515442e-06, 3.0199749e-06], [1.9813169e-09, 1.4367421e-06, 4.2192623e-06], [1.7577956e-09, 1.5941217e-06, 4.9128444e-06], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[1.8200443e-09, 1.3520319e-06, 3.0891476e-06], [1.6453449e-09, 1.5757826e-06, 4.3824580e-06], [1.3922893e-09, 1.7362590e-06, 4.9643909e-06], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[1.3885696e-09, 1.2316292e-06, 3.0375882e-06], [1.2445081e-09, 1.2671688e-06, 4.4312010e-06], [1.0393739e-09, 1.3993149e-06, 4.8259253e-06], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[6.6547938e-05, 6.5133048e-05, 5.4650016e-05], [6.6449466e-05, 6.5501496e-05, 5.4773409e-05], [6.6724380e-05, 6.6765897e-05, 5.5014516e-05], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[4.8890193e-05, 8.4378182e-05, 5.7880963e-05], [4.8967249e-05, 8.4766405e-05, 5.7902074e-05], [4.9593367e-05, 8.6330016e-05, 5.7977104e-05], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - QVRES(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg/s
- long_name :
- Rate of residual condensation term
- basename :
- QVRES
array([[[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., ... ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - RAINQM(time, lev, ncol)float326.281e-06 1.573e-07 ... 1.202e-24
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Grid box averaged rain amount
- basename :
- RAINQM
array([[[6.2806789e-06, 1.5730694e-07, 1.9481126e-07], [7.0419896e-06, 1.0044978e-07, 3.5804541e-07], [8.3821915e-06, 1.1156783e-07, 1.1010078e-06], ..., [1.1930321e-24, 1.1718242e-24, 1.1644136e-24], [1.2329296e-24, 1.2389521e-24, 1.2518271e-24], [1.2767012e-24, 1.2854699e-24, 1.2983939e-24]], [[4.9183936e-06, 4.3230539e-08, 2.3842873e-07], [5.5427658e-06, 1.9117327e-07, 4.2130219e-07], [6.6487332e-06, 1.0708184e-06, 1.3233270e-06], ..., [1.1880765e-24, 1.1644187e-24, 1.1603373e-24], [1.2509350e-24, 1.2738894e-24, 1.2872608e-24], [1.2633745e-24, 1.3343550e-24, 1.3554254e-24]], [[3.6210685e-07, 1.1235369e-07, 1.9843256e-07], [1.8983151e-06, 2.8573365e-07, 5.2020806e-07], [2.9729897e-06, 1.3607784e-06, 1.4758266e-06], ..., ... ..., [1.1142728e-24, 1.1179138e-24, 1.1178710e-24], [1.1602253e-24, 1.1616337e-24, 1.1588297e-24], [1.2266695e-24, 1.2541305e-24, 1.2672579e-24]], [[7.2427283e-06, 2.6780740e-06, 3.6680504e-08], [8.4554704e-06, 3.0667286e-06, 4.7371099e-08], [1.0819384e-05, 3.8294065e-06, 7.4915931e-08], ..., [1.1097641e-24, 1.1138300e-24, 1.1145571e-24], [1.1256490e-24, 1.1287824e-24, 1.1495750e-24], [1.1699555e-24, 1.2056228e-24, 1.2418144e-24]], [[7.9078045e-06, 5.2989767e-06, 1.5327441e-06], [8.9839041e-06, 5.9985491e-06, 1.8330389e-06], [1.0931299e-05, 7.3324632e-06, 2.4981409e-06], ..., [1.0875458e-24, 1.0983381e-24, 1.1067339e-24], [1.0989003e-24, 1.1301169e-24, 1.1648636e-24], [1.1318332e-24, 1.1649369e-24, 1.2020652e-24]]], dtype=float32) - RELHUM(time, lev, ncol)float3283.73 74.06 ... 0.0002164 0.0002172
- mdims :
- 9
- units :
- percent
- long_name :
- Relative humidity
- basename :
- RELHUM
array([[[8.3730804e+01, 7.4059944e+01, 8.2564720e+01], [8.5614693e+01, 7.5782059e+01, 8.3579903e+01], [8.8982658e+01, 7.8535530e+01, 8.6177773e+01], ..., [2.1726268e-04, 2.1736416e-04, 2.1712016e-04], [2.1540654e-04, 2.1414792e-04, 2.1438320e-04], [2.1281077e-04, 2.1633541e-04, 2.1900127e-04]], [[8.3065056e+01, 7.5226585e+01, 8.3333389e+01], [8.4900948e+01, 7.7023026e+01, 8.4370743e+01], [8.8228500e+01, 7.9907326e+01, 8.7039383e+01], ..., [2.1690367e-04, 2.1703786e-04, 2.1680453e-04], [2.1528378e-04, 2.1511430e-04, 2.1582222e-04], [2.1399544e-04, 2.1747973e-04, 2.2052214e-04]], [[8.2156151e+01, 7.6464828e+01, 8.4202568e+01], [8.3978065e+01, 7.8391991e+01, 8.5323051e+01], [8.7271988e+01, 8.1308601e+01, 8.8111702e+01], ..., ... ..., [2.1630268e-04, 2.1623749e-04, 2.1620565e-04], [2.1642138e-04, 2.1635329e-04, 2.1655482e-04], [2.1451162e-04, 2.1730708e-04, 2.1797139e-04]], [[8.5012337e+01, 8.7809311e+01, 8.9606087e+01], [8.6781754e+01, 8.9883835e+01, 9.1476593e+01], [8.9601952e+01, 9.2830826e+01, 9.4459862e+01], ..., [2.1639591e-04, 2.1618683e-04, 2.1608094e-04], [2.1660073e-04, 2.1666089e-04, 2.1642998e-04], [2.1596043e-04, 2.1666706e-04, 2.1773181e-04]], [[8.5153961e+01, 8.8320877e+01, 9.0189697e+01], [8.6953484e+01, 9.0313118e+01, 9.2181778e+01], [8.9808151e+01, 9.3310638e+01, 9.5297989e+01], ..., [2.1643350e-04, 2.1626646e-04, 2.1627167e-04], [2.1666149e-04, 2.1697833e-04, 2.1664714e-04], [2.1706083e-04, 2.1635288e-04, 2.1724954e-04]]], dtype=float32) - RERCLD(time, lev, ncol)float325.911e-05 3.797e-05 ... 0.0 0.0
- mdims :
- 9
- units :
- m
- long_name :
- Diagnostic effective radius of Liquid Cloud and Rain
- basename :
- RERCLD
array([[[5.91073149e-05, 3.79665726e-05, 4.17605625e-05], [5.19358910e-05, 4.14049064e-05, 4.36644768e-05], [4.40789590e-05, 4.52042659e-05, 4.66174897e-05], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[6.30436261e-05, 3.81821992e-05, 4.56247326e-05], [5.53605314e-05, 3.95952120e-05, 4.72612410e-05], [4.65019839e-05, 4.43411409e-05, 5.02099283e-05], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[9.77340314e-05, 4.61144846e-05, 4.55928348e-05], [7.24628189e-05, 4.99035159e-05, 4.82009709e-05], [5.03359188e-05, 5.62589230e-05, 5.31016485e-05], ..., ... ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[5.55629667e-05, 1.09471672e-04, 1.97550704e-04], [5.21807160e-05, 1.00631187e-04, 1.72877451e-04], [6.52848103e-05, 8.70505901e-05, 1.41538781e-04], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[5.49422730e-05, 6.70139270e-05, 1.17794450e-04], [5.06664546e-05, 7.73603460e-05, 1.00770529e-04], [6.11730356e-05, 9.24950655e-05, 7.77939204e-05], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]]], dtype=float32) - RHCFMIP(time, lev, ncol)float3287.35 76.8 ... 9.493e-06 8.739e-06
- mdims :
- 9
- units :
- percent
- long_name :
- Relative humidity with respect to water above 273 K, ice below 273 K
- basename :
- RHCFMIP
array([[[8.73468475e+01, 7.67996902e+01, 8.61570587e+01], [8.94625854e+01, 7.87407684e+01, 8.73249283e+01], [9.31331100e+01, 8.18148422e+01, 9.01377716e+01], ..., [3.14275967e-05, 2.79523865e-05, 2.56188632e-05], [1.22271085e-05, 8.69568976e-06, 7.92471565e-06], [8.82900440e-06, 6.15369618e-06, 5.53115069e-06]], [[8.66067123e+01, 7.79727097e+01, 8.69158554e+01], [8.86868210e+01, 7.99655685e+01, 8.81175690e+01], [9.22966232e+01, 8.32150497e+01, 9.10239258e+01], ..., [2.98583691e-05, 2.86222803e-05, 2.65356339e-05], [1.04015999e-05, 7.35037929e-06, 6.78888318e-06], [8.43694488e-06, 5.43379792e-06, 4.89474132e-06]], [[8.56140823e+01, 7.92143860e+01, 8.77677002e+01], [8.76887665e+01, 8.13232346e+01, 8.90711975e+01], [9.12698364e+01, 8.46393738e+01, 9.21332169e+01], ..., ... ..., [5.72765057e-05, 4.78244074e-05, 4.38797069e-05], [3.17296763e-05, 2.86792420e-05, 2.67102241e-05], [1.33338663e-05, 1.03805851e-05, 9.65839445e-06]], [[8.58085632e+01, 9.03497467e+01, 9.35536499e+01], [8.79123993e+01, 9.26797028e+01, 9.56211700e+01], [9.11677856e+01, 9.60781937e+01, 9.88732834e+01], ..., [5.63095200e-05, 4.78519214e-05, 4.40814838e-05], [2.83314002e-05, 2.42689530e-05, 2.27930286e-05], [1.30289027e-05, 1.00015177e-05, 9.64050832e-06]], [[8.60665512e+01, 9.05253143e+01, 9.38575058e+01], [8.81850281e+01, 9.28385086e+01, 9.60387344e+01], [9.14788971e+01, 9.62475891e+01, 9.95178833e+01], ..., [5.16498476e-05, 4.34627545e-05, 4.01654543e-05], [2.39641213e-05, 1.94484237e-05, 1.81505584e-05], [1.30828603e-05, 9.49347032e-06, 8.73908903e-06]]], dtype=float32) - RHI(time, lev, ncol)float3287.35 76.8 ... 9.493e-06 8.739e-06
- mdims :
- 9
- units :
- percent
- long_name :
- Relative humidity with respect to ice
- basename :
- RHI
array([[[8.73468475e+01, 7.67996902e+01, 8.61570587e+01], [8.94625854e+01, 7.87407684e+01, 8.73249283e+01], [9.31331100e+01, 8.18148422e+01, 9.01377716e+01], ..., [3.14275967e-05, 2.79523865e-05, 2.56188632e-05], [1.22271085e-05, 8.69568976e-06, 7.92471565e-06], [8.82900440e-06, 6.15369618e-06, 5.53115069e-06]], [[8.66067123e+01, 7.79727097e+01, 8.69158554e+01], [8.86868210e+01, 7.99655685e+01, 8.81175690e+01], [9.22966232e+01, 8.32150497e+01, 9.10239258e+01], ..., [2.98583691e-05, 2.86222803e-05, 2.65356339e-05], [1.04015999e-05, 7.35037929e-06, 6.78888318e-06], [8.43694488e-06, 5.43379792e-06, 4.89474132e-06]], [[8.56140823e+01, 7.92143860e+01, 8.77677002e+01], [8.76887665e+01, 8.13232346e+01, 8.90711975e+01], [9.12698364e+01, 8.46393738e+01, 9.21332169e+01], ..., ... ..., [5.72765057e-05, 4.78244074e-05, 4.38797069e-05], [3.17296763e-05, 2.86792420e-05, 2.67102241e-05], [1.33338663e-05, 1.03805851e-05, 9.65839445e-06]], [[8.58085632e+01, 9.03497467e+01, 9.35536499e+01], [8.79123993e+01, 9.26797028e+01, 9.56211700e+01], [9.11677856e+01, 9.60781937e+01, 9.88732834e+01], ..., [5.63095200e-05, 4.78519214e-05, 4.40814838e-05], [2.83314002e-05, 2.42689530e-05, 2.27930286e-05], [1.30289027e-05, 1.00015177e-05, 9.64050832e-06]], [[8.60665512e+01, 9.05253143e+01, 9.38575058e+01], [8.81850281e+01, 9.28385086e+01, 9.60387344e+01], [9.14788971e+01, 9.62475891e+01, 9.95178833e+01], ..., [5.16498476e-05, 4.34627545e-05, 4.01654543e-05], [2.39641213e-05, 1.94484237e-05, 1.81505584e-05], [1.30828603e-05, 9.49347032e-06, 8.73908903e-06]]], dtype=float32) - RHW(time, lev, ncol)float3282.26 73.25 ... 0.0002164 0.0002172
- mdims :
- 9
- units :
- percent
- long_name :
- Relative humidity with respect to liquid
- basename :
- RHW
array([[[8.2255363e+01, 7.3248878e+01, 8.1090027e+01], [8.3940842e+01, 7.4830933e+01, 8.1965187e+01], [8.6876579e+01, 7.7299622e+01, 8.4184753e+01], ..., [2.1726268e-04, 2.1736416e-04, 2.1712016e-04], [2.1540654e-04, 2.1414792e-04, 2.1438320e-04], [2.1281077e-04, 2.1633541e-04, 2.1900127e-04]], [[8.1629616e+01, 7.4459305e+01, 8.1875824e+01], [8.3288261e+01, 7.6087341e+01, 8.2788376e+01], [8.6178467e+01, 7.8714394e+01, 8.5099106e+01], ..., [2.1690367e-04, 2.1703786e-04, 2.1680453e-04], [2.1528378e-04, 2.1511430e-04, 2.1582222e-04], [2.1399544e-04, 2.1747973e-04, 2.2052214e-04]], [[8.0763603e+01, 7.5732140e+01, 8.2760506e+01], [8.2425003e+01, 7.7464752e+01, 8.3767723e+01], [8.5300423e+01, 8.0150711e+01, 8.6221703e+01], ..., ... ..., [2.1630268e-04, 2.1623749e-04, 2.1620565e-04], [2.1642138e-04, 2.1635329e-04, 2.1655482e-04], [2.1451162e-04, 2.1730708e-04, 2.1797139e-04]], [[8.4971191e+01, 8.7355415e+01, 8.7961464e+01], [8.6743958e+01, 8.9279167e+01, 8.9617226e+01], [8.9444786e+01, 9.2013275e+01, 9.2217079e+01], ..., [2.1639591e-04, 2.1618683e-04, 2.1608094e-04], [2.1660073e-04, 2.1666089e-04, 2.1642998e-04], [2.1596043e-04, 2.1666706e-04, 2.1773181e-04]], [[8.5120422e+01, 8.7962250e+01, 8.8939125e+01], [8.6903458e+01, 8.9879257e+01, 9.0708588e+01], [8.9634750e+01, 9.2640251e+01, 9.3525269e+01], ..., [2.1643350e-04, 2.1626646e-04, 2.1627167e-04], [2.1666149e-04, 2.1697833e-04, 2.1664714e-04], [2.1706083e-04, 2.1635288e-04, 2.1724954e-04]]], dtype=float32) - SNOWQM(time, lev, ncol)float322.413e-09 1.036e-06 ... 2.229e-24
- mdims :
- 9
- units :
- kg/kg
- long_name :
- Grid box averaged snow amount
- basename :
- SNOWQM
array([[[2.4134366e-09, 1.0363789e-06, 2.5305048e-06], [2.2085680e-09, 1.0515073e-06, 4.3549912e-06], [1.8306943e-09, 1.6368321e-06, 4.9030259e-06], ..., [2.5175845e-24, 2.5078636e-24, 2.4833471e-24], [2.2932725e-24, 2.2098175e-24, 2.2110994e-24], [2.2278446e-24, 2.1949179e-24, 2.2116449e-24]], [[2.0164521e-09, 1.1130018e-06, 2.6136734e-06], [1.8323438e-09, 1.1368479e-06, 4.5311467e-06], [1.4693483e-09, 1.9019867e-06, 4.9401456e-06], ..., [2.4975058e-24, 2.5029738e-24, 2.4886000e-24], [2.3096059e-24, 2.2448944e-24, 2.2506959e-24], [2.2531705e-24, 2.2221062e-24, 2.2304078e-24]], [[1.6153280e-09, 8.1774061e-07, 2.4883022e-06], [1.4304888e-09, 1.3079732e-06, 4.4635240e-06], [1.0998460e-09, 1.4218654e-06, 4.6559280e-06], ..., ... ..., [2.4879665e-24, 2.4637232e-24, 2.4576474e-24], [2.3235373e-24, 2.3143242e-24, 2.3109761e-24], [2.1786043e-24, 2.1922802e-24, 2.1949836e-24]], [[6.7375637e-05, 6.6274610e-05, 5.4352276e-05], [6.6785564e-05, 6.6455323e-05, 5.4327065e-05], [6.5703716e-05, 6.6877437e-05, 5.4217737e-05], ..., [2.4952313e-24, 2.4836544e-24, 2.4732539e-24], [2.3196021e-24, 2.3171271e-24, 2.3043828e-24], [2.2145613e-24, 2.2058180e-24, 2.2061412e-24]], [[5.0289938e-05, 8.5391461e-05, 5.9973376e-05], [4.9895767e-05, 8.5398839e-05, 6.0003069e-05], [4.9214934e-05, 8.5479660e-05, 6.0095492e-05], ..., [2.4910342e-24, 2.4823743e-24, 2.4755895e-24], [2.3228197e-24, 2.3078031e-24, 2.2795694e-24], [2.2374886e-24, 2.2231325e-24, 2.2287567e-24]]], dtype=float32) - SNOW_ICLD_VISTAU(time, lev, ncol)float320.0 0.001852 0.005498 ... 0.0 0.0
- mdims :
- 9
- Sampling_Sequence :
- rad_lwsw
- units :
- 1
- long_name :
- Snow in-cloud extinction visible sw optical depth
- basename :
- SNOW_ICLD_VISTAU
array([[[0. , 0.00185158, 0.00549758], [0. , 0.00457868, 0.01790487], [0. , 0.00762325, 0.02894233], ..., [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ]], [[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan]], [[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., ... ..., [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ]], [[0.04121059, 0.02919545, 0.02661125], [0.09546688, 0.06604153, 0.05730582], [0.14410475, 0.09598774, 0.07661083], ..., [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ]], [[0.03478727, 0.05341719, 0.02438943], [0.07941503, 0.1212134 , 0.05238835], [0.11783466, 0.1780459 , 0.0696812 ], ..., [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ]]], dtype=float32) - SOLIN(time, ncol)float322.817 15.63 30.87 ... 64.11 83.85
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Solar insolation
- basename :
- SOLIN
array([[2.81735039e+00, 1.56292133e+01, 3.08679790e+01], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 5.35354950e-03], [1.90454841e+00, 2.76260605e+01, 3.71853752e+01], [3.53002853e+01, 7.89947128e+01, 9.28021622e+01], [6.08145294e+01, 1.03142754e+02, 1.20023964e+02], [5.97802429e+01, 9.80733414e+01, 1.16977371e+02], [3.22491226e+01, 6.41135635e+01, 8.38517838e+01]], dtype=float32) - SWCF(time, ncol)float32-0.3587 -1.876 ... -14.44 -12.42
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Shortwave cloud forcing
- basename :
- SWCF
array([[-3.5865873e-01, -1.8764081e+00, -2.2877464e+00], [ 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [ 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [ 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [ 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [ 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [ 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [ 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [ 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [ 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [ 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [ 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [ 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [ 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [ 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [ 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [ 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [ 0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [ 0.0000000e+00, 0.0000000e+00, 6.4974687e-05], [ 2.7761202e-02, -1.6292611e+00, -4.4913950e+00], [-8.6251812e+00, -1.5767940e+01, -1.2997323e+01], [-1.8027096e+01, -2.3565535e+01, -1.7638680e+01], [-1.7337543e+01, -2.4123243e+01, -1.8091593e+01], [-7.9928007e+00, -1.4435503e+01, -1.2423702e+01]], dtype=float32) - T(time, lev, ncol)float32267.0 268.3 266.9 ... 264.1 265.1
- mdims :
- 9
- units :
- K
- long_name :
- Temperature
- basename :
- T
array([[[266.9897 , 268.29642, 266.93283], [266.61496, 267.9273 , 266.65262], [266.0163 , 267.3274 , 266.14108], ..., [259.49014, 260.7848 , 261.74 ], [265.60776, 269.5085 , 270.6233 ], [264.73633, 269.11035, 270.51874]], [[267.0793 , 268.423 , 267.0227 ], [266.70782, 268.0522 , 266.75085], [266.1142 , 267.4473 , 266.2457 ], ..., [260.034 , 260.50656, 261.3327 ], [267.46976, 271.5637 , 272.5604 ], [265.3187 , 270.64722, 272.0655 ]], [[267.16782, 268.5413 , 267.1248 ], [266.80008, 268.16577, 266.85312], [266.2113 , 267.56146, 266.34753], ..., ... ..., [253.03531, 254.92351, 255.83473], [255.15038, 256.2214 , 256.99286], [260.20685, 263.13892, 263.9881 ]], [[272.16 , 269.69852, 266.82767], [271.79153, 269.3201 , 266.4981 ], [271.2041 , 268.71872, 266.00067], ..., [253.21716, 254.91496, 255.77979], [256.36392, 258.0316 , 258.70105], [260.53613, 263.5252 , 263.99664]], [[272.03137, 270.21118, 267.6298 ], [271.66174, 269.8332 , 267.29352], [271.07172, 269.23587, 266.77948], ..., [254.12259, 255.93938, 256.78268], [258.16855, 260.46756, 261.2148 ], [260.5466 , 264.09888, 265.0888 ]]], dtype=float32) - TGCLDCWP(time, ncol)float320.1811 0.2282 ... 0.2024 0.1926
- units :
- kg/m2
- long_name :
- Total grid-box cloud water path (liquid and ice)
- basename :
- TGCLDCWP
array([[0.18114646, 0.22820754, 0.15575309], [0.1744934 , 0.22448756, 0.1601221 ], [0.1680441 , 0.2137294 , 0.1587637 ], [0.16202153, 0.2066004 , 0.14971939], [0.15008987, 0.20008753, 0.13325696], [0.13618869, 0.19012673, 0.11910434], [0.11910637, 0.17518578, 0.102689 ], [0.10712057, 0.15127365, 0.08371527], [0.09396417, 0.1167035 , 0.06592898], [0.08553578, 0.07963008, 0.04692875], [0.10250235, 0.04724938, 0.02032844], [0.1205819 , 0.02366849, 0.00811968], [0.15363166, 0.01728513, 0.0046124 ], [0.1826474 , 0.0149369 , 0.00580784], [0.20299254, 0.00957081, 0.0065171 ], [0.22154106, 0.01748713, 0.00639987], [0.22008835, 0.03790848, 0.01777298], [0.22203597, 0.04624197, 0.03639105], [0.18396242, 0.05716376, 0.04453827], [0.18612462, 0.05571092, 0.05654092], [0.2359936 , 0.07914568, 0.06788007], [0.27899826, 0.13047609, 0.09561998], [0.31076473, 0.18815896, 0.1481685 ], [0.32284176, 0.20238695, 0.19256572]], dtype=float32) - TGCLDIWP(time, ncol)float323.246e-07 0.00132 ... 0.01427
- units :
- kg/m2
- long_name :
- Total grid-box cloud ice water path
- basename :
- TGCLDIWP
array([[3.24580526e-07, 1.31966267e-03, 4.42083320e-03], [1.12135123e-04, 1.13189640e-03, 3.59549257e-03], [3.06180649e-04, 1.05145539e-03, 3.12791532e-03], [5.21361304e-04, 1.43216643e-03, 3.33235646e-03], [7.94848427e-04, 1.13456463e-03, 3.65875405e-03], [2.78199324e-03, 1.03811361e-03, 3.23465979e-03], [5.08557213e-03, 1.27887225e-03, 2.96267611e-03], [6.82581076e-03, 1.79685338e-03, 2.82059936e-03], [8.12589191e-03, 2.25723512e-03, 1.87052554e-03], [8.12869892e-03, 2.90191313e-03, 1.52494211e-03], [8.25674646e-03, 3.42579628e-03, 1.44140783e-03], [9.56819020e-03, 4.76081250e-03, 2.56005512e-03], [9.55020078e-03, 6.15129434e-03, 4.61240346e-03], [9.77243297e-03, 7.22850347e-03, 5.80784213e-03], [1.47658149e-02, 7.06784567e-03, 6.51709549e-03], [1.75385252e-02, 7.96106923e-03, 6.39986992e-03], [1.53730633e-02, 9.86036938e-03, 8.33041128e-03], [4.01215740e-02, 2.26726923e-02, 1.38317840e-02], [2.60271616e-02, 3.59481648e-02, 2.32495293e-02], [1.51887257e-02, 2.59178039e-02, 2.61951741e-02], [1.15926182e-02, 2.19484530e-02, 1.96199566e-02], [1.20477751e-02, 1.66240074e-02, 1.49319135e-02], [1.13811819e-02, 2.04702448e-02, 1.68968718e-02], [1.46324420e-02, 1.81456879e-02, 1.42741315e-02]], dtype=float32) - TGCLDLWP(time, ncol)float320.1811 0.2269 ... 0.1842 0.1783
- units :
- kg/m2
- long_name :
- Total grid-box cloud liquid water path
- basename :
- TGCLDLWP
array([[1.81146145e-01, 2.26887882e-01, 1.51332259e-01], [1.74381271e-01, 2.23355666e-01, 1.56526595e-01], [1.67737916e-01, 2.12677941e-01, 1.55635789e-01], [1.61500171e-01, 2.05168232e-01, 1.46387026e-01], [1.49295032e-01, 1.98952973e-01, 1.29598200e-01], [1.33406684e-01, 1.89088613e-01, 1.15869679e-01], [1.14020795e-01, 1.73906907e-01, 9.97263268e-02], [1.00294754e-01, 1.49476796e-01, 8.08946639e-02], [8.58382806e-02, 1.14446267e-01, 6.40584528e-02], [7.74070844e-02, 7.67281651e-02, 4.54038046e-02], [9.42456052e-02, 4.38235849e-02, 1.88870281e-02], [1.11013710e-01, 1.89076792e-02, 5.55962836e-03], [1.44081458e-01, 1.11338329e-02, 9.72856846e-14], [1.72874972e-01, 7.70840049e-03, 0.00000000e+00], [1.88226730e-01, 2.50296178e-03, 0.00000000e+00], [2.04002544e-01, 9.52606648e-03, 0.00000000e+00], [2.04715282e-01, 2.80481130e-02, 9.44256876e-03], [1.81914404e-01, 2.35692784e-02, 2.25592665e-02], [1.57935262e-01, 2.12155953e-02, 2.12887377e-02], [1.70935899e-01, 2.97931191e-02, 3.03457435e-02], [2.24400982e-01, 5.71972243e-02, 4.82601188e-02], [2.66950458e-01, 1.13852076e-01, 8.06880593e-02], [2.99383551e-01, 1.67688712e-01, 1.31271631e-01], [3.08209330e-01, 1.84241250e-01, 1.78291589e-01]], dtype=float32) - TOT_CLD_VISTAU(time, lev, ncol)float320.0 0.0004629 0.001374 ... 0.0 0.0
- mdims :
- 9
- Sampling_Sequence :
- rad_lwsw
- units :
- 1
- long_name :
- Total gbx cloud extinction visible sw optical depth
- basename :
- TOT_CLD_VISTAU
array([[[0. , 0.00046289, 0.00137439], [0. , 0.00114467, 0.00447622], [0. , 0.00190581, 0.00723679], ..., [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ]], [[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan]], [[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., ... ..., [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ]], [[0.01030265, 0.00729886, 0.00665281], [0.02386672, 0.01651038, 0.01432646], [0.03602619, 0.02399693, 0.01915271], ..., [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ]], [[0.00869682, 0.0133543 , 0.00609736], [0.01985376, 0.03030335, 0.01309709], [0.02945866, 0.04451147, 0.0174203 ], ..., [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ]]], dtype=float32) - TOT_ICLD_VISTAU(time, lev, ncol)float320.0 0.001852 0.005498 ... 0.0 0.0
- mdims :
- 9
- Sampling_Sequence :
- rad_lwsw
- units :
- 1
- long_name :
- Total in-cloud extinction visible sw optical depth
- basename :
- TOT_ICLD_VISTAU
array([[[0. , 0.00185158, 0.00549758], [0. , 0.00457868, 0.01790487], [0. , 0.00762325, 0.02894717], ..., [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ]], [[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., [ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan]], [[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., ... ..., [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ]], [[0.04121059, 0.02919545, 0.02661125], [0.09546688, 0.06604153, 0.05730582], [0.14410475, 0.09598774, 0.07661083], ..., [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ]], [[0.03478727, 0.05341719, 0.02438943], [0.07941503, 0.1212134 , 0.05238835], [0.11783466, 0.1780459 , 0.0696812 ], ..., [0. , 0. , 0. ], [0. , 0. , 0. ], [0. , 0. , 0. ]]], dtype=float32) - U(time, lev, ncol)float320.2622 -0.7117 ... 98.55 105.0
- mdims :
- 9
- units :
- m/s
- long_name :
- Zonal wind
- basename :
- U
array([[[ 2.62227178e-01, -7.11740911e-01, -1.63492262e-01], [ 2.73156792e-01, -7.89959013e-01, -2.96294153e-01], [ 2.67408490e-01, -8.66507173e-01, -4.78364408e-01], ..., [ 5.03317871e+01, 4.32601776e+01, 4.04384232e+01], [ 6.17892380e+01, 5.82935257e+01, 5.91240730e+01], [ 5.88163261e+01, 4.24368095e+01, 3.91790047e+01]], [[ 5.33694029e-01, -6.04423523e-01, -7.19045550e-02], [ 5.70249259e-01, -6.74651861e-01, -1.80030644e-01], [ 5.82710385e-01, -7.46855438e-01, -3.66099358e-01], ..., [ 5.19736214e+01, 4.46206360e+01, 4.21126404e+01], [ 6.80857620e+01, 6.81021881e+01, 7.06165543e+01], [ 5.86999359e+01, 4.38686447e+01, 4.06282654e+01]], [[ 8.42299521e-01, -4.85764980e-01, 1.08472900e-02], [ 9.08994913e-01, -5.46166718e-01, -7.60713965e-02], [ 9.41705227e-01, -6.11726642e-01, -2.69435018e-01], ..., ... [ 4.76525192e+01, 5.33234062e+01, 5.57136307e+01], [ 5.41735802e+01, 6.50180435e+01, 7.05370712e+01], [ 6.53356934e+01, 8.18249435e+01, 8.95381622e+01]], [[ 1.10706186e+01, 1.06644993e+01, 7.90016890e+00], [ 1.21658077e+01, 1.24800549e+01, 1.05907450e+01], [ 1.28817434e+01, 1.38602705e+01, 1.27675409e+01], ..., [ 5.03022385e+01, 5.75731888e+01, 5.97751389e+01], [ 6.22654305e+01, 7.58059082e+01, 8.07216263e+01], [ 7.21504669e+01, 9.01455536e+01, 9.70511627e+01]], [[ 1.04807138e+01, 1.09754581e+01, 8.52511120e+00], [ 1.14669409e+01, 1.28784132e+01, 1.12409277e+01], [ 1.20999355e+01, 1.43055105e+01, 1.33642635e+01], ..., [ 5.69905548e+01, 6.49683990e+01, 6.63153534e+01], [ 7.20876846e+01, 8.73528824e+01, 9.15143890e+01], [ 7.87675095e+01, 9.85469666e+01, 1.04994057e+02]]], dtype=float32) - V(time, lev, ncol)float32-4.542 -6.798 ... 13.94 17.55
- mdims :
- 9
- units :
- m/s
- long_name :
- Meridional wind
- basename :
- V
array([[[-4.542304 , -6.798095 , -4.8485556 ], [-4.8645105 , -7.3514667 , -6.721163 ], [-4.9855566 , -7.6808047 , -7.652571 ], ..., [ 8.1667385 , 6.158981 , 6.565375 ], [17.39923 , 9.301125 , 7.9279423 ], [-4.427489 , -4.0905447 , -2.853115 ]], [[-3.9623654 , -6.6235547 , -4.723304 ], [-4.2424593 , -7.1672573 , -6.615004 ], [-4.339403 , -7.489289 , -7.562458 ], ..., [ 9.337824 , 7.486973 , 7.132428 ], [15.605526 , 7.6830273 , 6.1184096 ], [-3.1699805 , -5.099699 , -4.4647074 ]], [[-3.2448413 , -6.351094 , -4.554724 ], [-3.4740312 , -6.8776445 , -6.4175963 ], [-3.54404 , -7.188098 , -7.3535576 ], ..., ... ..., [ 2.726963 , -1.4200326 , -1.9484863 ], [11.470686 , 5.9823194 , 5.452907 ], [11.887689 , 5.2791543 , 5.499318 ]], [[ 0.1323834 , 7.7495565 , 6.7964725 ], [ 0.11003168, 8.950782 , 8.894286 ], [ 0.03844166, 9.675483 , 10.129004 ], ..., [ 8.446177 , 4.7143297 , 3.9297893 ], [13.59661 , 9.956541 , 10.788858 ], [13.364609 , 10.260371 , 12.646693 ]], [[-2.196862 , 6.865246 , 6.369951 ], [-2.4374077 , 7.9396367 , 8.215149 ], [-2.6460738 , 8.556952 , 9.270718 ], ..., [13.065639 , 9.320509 , 8.591337 ], [13.801663 , 11.897252 , 13.399579 ], [13.535832 , 13.939431 , 17.549086 ]]], dtype=float32) - WSUB(time, lev, ncol)float320.2996 0.4065 0.3273 ... 0.2 0.2
- mdims :
- 9
- units :
- m/s
- long_name :
- Diagnostic sub-grid vertical velocity
- basename :
- WSUB
array([[[0.2996484 , 0.4065365 , 0.32726038], [0.40532094, 0.45577952, 0.3072182 ], [0.49058613, 0.49311602, 0.26186675], ..., [0.2 , 0.2 , 0.2 ], [0.2 , 0.2 , 0.2 ], [0.2 , 0.2 , 0.2 ]], [[0.27994096, 0.39706534, 0.31210077], [0.3851581 , 0.44357398, 0.2919714 ], [0.46935824, 0.4771005 , 0.24735034], ..., [0.2 , 0.2 , 0.2 ], [0.2 , 0.2 , 0.2 ], [0.2 , 0.2 , 0.2 ]], [[0.2582274 , 0.38286006, 0.2987275 ], [0.36366802, 0.42694718, 0.2793207 ], [0.44693393, 0.4574265 , 0.23638323], ..., ... ..., [0.2 , 0.2 , 0.2 ], [0.2 , 0.2 , 0.2 ], [0.2 , 0.2 , 0.2 ]], [[0.54244083, 0.7869016 , 0.67820954], [0.5462084 , 0.7931695 , 0.64544046], [0.524783 , 0.7426842 , 0.5395676 ], ..., [0.2 , 0.2 , 0.2 ], [0.2 , 0.2 , 0.2 ], [0.2 , 0.2 , 0.2 ]], [[0.5149096 , 0.7755813 , 0.70120794], [0.52190924, 0.77981424, 0.67150956], [0.50700366, 0.72882956, 0.5715366 ], ..., [0.2 , 0.2 , 0.2 ], [0.2 , 0.2 , 0.2 ], [0.2 , 0.2 , 0.2 ]]], dtype=float32) - WSUBI(time, lev, ncol)float320.2996 0.4065 ... 0.001 0.001
- mdims :
- 9
- units :
- m/s
- long_name :
- Diagnostic sub-grid vertical velocity for ice
- basename :
- WSUBI
array([[[0.2996484 , 0.4065365 , 0.32726038], [0.40532094, 0.45577952, 0.3072182 ], [0.49058613, 0.49311602, 0.26186675], ..., [0.001 , 0.001 , 0.001 ], [0.001 , 0.001 , 0.001 ], [0.001 , 0.001 , 0.001 ]], [[0.27994096, 0.39706534, 0.31210077], [0.3851581 , 0.44357398, 0.2919714 ], [0.46935824, 0.4771005 , 0.24735034], ..., [0.001 , 0.001 , 0.001 ], [0.001 , 0.001 , 0.001 ], [0.001 , 0.001 , 0.001 ]], [[0.2582274 , 0.38286006, 0.2987275 ], [0.36366802, 0.42694718, 0.2793207 ], [0.44693393, 0.4574265 , 0.23638323], ..., ... ..., [0.001 , 0.001 , 0.001 ], [0.001 , 0.001 , 0.001 ], [0.001 , 0.001 , 0.001 ]], [[0.54244083, 0.7869016 , 0.67820954], [0.5462084 , 0.7931695 , 0.64544046], [0.524783 , 0.7426842 , 0.5395676 ], ..., [0.001 , 0.001 , 0.001 ], [0.001 , 0.001 , 0.001 ], [0.001 , 0.001 , 0.001 ]], [[0.5149096 , 0.7755813 , 0.70120794], [0.52190924, 0.77981424, 0.67150956], [0.50700366, 0.72882956, 0.5715366 ], ..., [0.001 , 0.001 , 0.001 ], [0.001 , 0.001 , 0.001 ], [0.001 , 0.001 , 0.001 ]]], dtype=float32) - Z3(time, lev, ncol)float3211.83 32.15 ... 6.079e+04 6.09e+04
- mdims :
- 9
- units :
- m
- long_name :
- Geopotential Height (above sea level)
- basename :
- Z3
array([[[1.18272314e+01, 3.21489334e+01, 9.00170975e+01], [4.90513115e+01, 6.95554352e+01, 1.27241875e+02], [1.08704430e+02, 1.29490067e+02, 1.86882751e+02], ..., [5.51122305e+04, 5.52486680e+04, 5.52995977e+04], [5.80696875e+04, 5.82353867e+04, 5.82979727e+04], [6.10566914e+04, 6.12689961e+04, 6.13457930e+04]], [[1.18311729e+01, 3.21548691e+01, 9.00212860e+01], [4.90680275e+01, 6.95799866e+01, 1.27260124e+02], [1.08743370e+02, 1.29545151e+02, 1.86926559e+02], ..., [5.50763984e+04, 5.52209297e+04, 5.52774688e+04], [5.80474062e+04, 5.82176562e+04, 5.82844648e+04], [6.10481758e+04, 6.12714961e+04, 6.13519062e+04]], [[1.18350210e+01, 3.21604462e+01, 9.00260391e+01], [4.90844231e+01, 6.96027756e+01, 1.27279961e+02], [1.08781670e+02, 1.29596863e+02, 1.86972305e+02], ..., ... ..., [5.47203359e+04, 5.49031875e+04, 5.49893828e+04], [5.75825391e+04, 5.77820586e+04, 5.78777305e+04], [6.04851328e+04, 6.07072031e+04, 6.08120000e+04]], [[1.20629940e+01, 3.22153587e+01, 9.00135422e+01], [5.00332642e+01, 6.98310242e+01, 1.27222481e+02], [1.10843285e+02, 1.30082382e+02, 1.86825974e+02], ..., [5.47320078e+04, 5.49215078e+04, 5.50064805e+04], [5.76020703e+04, 5.78105234e+04, 5.79041406e+04], [6.05133555e+04, 6.07480352e+04, 6.08480781e+04]], [[1.20571594e+01, 3.22388611e+01, 9.00500641e+01], [5.00088692e+01, 6.99288788e+01, 1.27373878e+02], [1.10787292e+02, 1.30296249e+02, 1.87155716e+02], ..., [5.47256758e+04, 5.49285156e+04, 5.50177266e+04], [5.76110000e+04, 5.78370234e+04, 5.79351914e+04], [6.05325078e+04, 6.07914844e+04, 6.08994375e+04]]], dtype=float32) - bc_a1(time, lev, ncol)float321.782e-12 1.903e-12 ... 4.415e-22
- mdims :
- 9
- units :
- kg/kg
- long_name :
- bc_a1 concentration
- basename :
- bc_a1
array([[[1.78161143e-12, 1.90279650e-12, 2.23142186e-12], [1.78400025e-12, 1.90467629e-12, 2.22964962e-12], [1.78832741e-12, 1.90788813e-12, 2.22263267e-12], ..., [1.47248440e-20, 1.16415695e-20, 1.08504431e-20], [1.73021445e-21, 1.57091120e-21, 1.41776010e-21], [1.13193740e-21, 1.50680672e-21, 1.54067165e-21]], [[1.83297626e-12, 1.85809744e-12, 2.23162808e-12], [1.83471120e-12, 1.86003600e-12, 2.22999787e-12], [1.83787512e-12, 1.86292301e-12, 2.22228204e-12], ..., [1.47088449e-20, 1.26281084e-20, 1.18266342e-20], [1.56642108e-21, 1.12979997e-21, 1.06001681e-21], [1.18879072e-21, 1.03587721e-21, 1.10892760e-21]], [[1.94037625e-12, 1.82106175e-12, 2.23691963e-12], [1.94191603e-12, 1.82268653e-12, 2.23521960e-12], [1.94469896e-12, 1.82451645e-12, 2.22511332e-12], ..., ... ..., [1.27967135e-20, 9.99291291e-21, 9.25688292e-21], [1.30818533e-21, 1.23629342e-21, 1.14959950e-21], [5.54385494e-22, 4.42887616e-22, 4.23573070e-22]], [[3.69944581e-13, 2.50064899e-12, 3.14817139e-12], [3.70134289e-13, 2.49993667e-12, 3.13959123e-12], [3.70477791e-13, 2.49800766e-12, 3.11334855e-12], ..., [1.39930008e-20, 1.20425081e-20, 1.14324301e-20], [1.04740998e-21, 1.01813221e-21, 1.10965471e-21], [4.82775503e-22, 4.16519921e-22, 4.28098127e-22]], [[3.20055180e-13, 2.07893620e-12, 2.57206778e-12], [3.20379113e-13, 2.07641695e-12, 2.55743213e-12], [3.21009739e-13, 2.06984907e-12, 2.51375548e-12], ..., [1.42533010e-20, 1.28880491e-20, 1.20271463e-20], [8.49511258e-22, 9.13060782e-22, 9.90419335e-22], [4.43285454e-22, 4.16090276e-22, 4.41451662e-22]]], dtype=float32) - bc_a3(time, lev, ncol)float327.966e-14 2.719e-13 ... 1.005e-36
- mdims :
- 9
- units :
- kg/kg
- long_name :
- bc_a3 concentration
- basename :
- bc_a3
array([[[7.96646413e-14, 2.71886842e-13, 3.86311290e-13], [8.09841289e-14, 2.71065641e-13, 4.00474548e-13], [8.66015904e-14, 2.69299584e-13, 4.08427632e-13], ..., [1.00000004e-36, 1.00012559e-36, 1.00008766e-36], [1.00050558e-36, 1.00000004e-36, 1.00000004e-36], [1.00000004e-36, 1.00814192e-36, 1.01708601e-36]], [[7.78791975e-14, 3.37308089e-13, 4.38303067e-13], [7.89440805e-14, 3.35871088e-13, 4.57460187e-13], [8.38406222e-14, 3.56054324e-13, 4.74962192e-13], ..., [1.00003950e-36, 1.00013797e-36, 1.00000004e-36], [1.00019653e-36, 1.00000004e-36, 1.00000004e-36], [1.00000004e-36, 1.00638619e-36, 1.02306483e-36]], [[1.82066981e-13, 3.16041925e-13, 4.83173126e-13], [1.94093588e-13, 3.85162090e-13, 5.06835622e-13], [1.93940770e-13, 4.37559846e-13, 5.14927348e-13], ..., ... ..., [1.00000004e-36, 1.00000004e-36, 1.00000004e-36], [1.00029940e-36, 1.00000004e-36, 1.00000004e-36], [1.00000004e-36, 1.00746203e-36, 1.00878010e-36]], [[3.82411184e-14, 7.06400948e-14, 8.50353723e-14], [3.86065759e-14, 7.09113351e-14, 8.47215297e-14], [3.83515377e-14, 7.12349762e-14, 8.38113216e-14], ..., [1.00003250e-36, 1.00000004e-36, 1.00000004e-36], [1.00014326e-36, 1.00006524e-36, 1.00000004e-36], [1.00229852e-36, 1.00456930e-36, 1.00836855e-36]], [[5.03119919e-14, 5.48661729e-14, 6.37070353e-14], [5.09702686e-14, 5.48437740e-14, 6.30605594e-14], [5.07503382e-14, 5.45964234e-14, 6.13599205e-14], ..., [1.00000004e-36, 1.00007663e-36, 1.00000004e-36], [1.00000004e-36, 1.00063652e-36, 1.00000004e-36], [1.00562415e-36, 1.00114215e-36, 1.00483189e-36]]], dtype=float32) - bc_a4(time, lev, ncol)float322.008e-12 4.952e-12 ... 1.327e-18
- mdims :
- 9
- units :
- kg/kg
- long_name :
- bc_a4 concentration
- basename :
- bc_a4
array([[[2.0082790e-12, 4.9516294e-12, 8.2505192e-12], [2.0099554e-12, 4.9349756e-12, 8.0790903e-12], [2.0134575e-12, 4.9003462e-12, 7.6998356e-12], ..., [2.2444537e-18, 2.2032731e-18, 2.1509006e-18], [1.7253130e-18, 1.6273522e-18, 1.6364575e-18], [1.6471388e-18, 1.5858806e-18, 1.5795480e-18]], [[2.0631329e-12, 5.2327934e-12, 8.9090757e-12], [2.0646002e-12, 5.2148390e-12, 8.6887329e-12], [2.0676362e-12, 5.1778508e-12, 8.1977931e-12], ..., [2.2291471e-18, 2.1630496e-18, 2.1284058e-18], [1.8015125e-18, 1.7001601e-18, 1.7175168e-18], [1.6567445e-18, 1.6558055e-18, 1.6600101e-18]], [[2.1179243e-12, 5.5110534e-12, 9.4779228e-12], [2.1191419e-12, 5.4919485e-12, 9.2152674e-12], [2.1216546e-12, 5.4528218e-12, 8.6372671e-12], ..., ... ..., [1.8736445e-18, 1.8304345e-18, 1.8220442e-18], [1.7274314e-18, 1.7240147e-18, 1.7020915e-18], [1.5063759e-18, 1.4756741e-18, 1.4774114e-18]], [[5.7437686e-13, 5.1017936e-12, 7.2563140e-12], [5.7369306e-13, 5.0859581e-12, 7.2064403e-12], [5.7223188e-13, 5.0481780e-12, 7.0588396e-12], ..., [1.8683528e-18, 1.8648020e-18, 1.8731581e-18], [1.5567542e-18, 1.5685327e-18, 1.6343458e-18], [1.2989582e-18, 1.3227084e-18, 1.4112126e-18]], [[6.4699591e-13, 4.8542585e-12, 6.1419407e-12], [6.4660609e-13, 4.8387019e-12, 6.0789390e-12], [6.4586184e-13, 4.8015650e-12, 5.8944707e-12], ..., [1.8805480e-18, 1.9087110e-18, 1.9198819e-18], [1.4065622e-18, 1.4745890e-18, 1.5715146e-18], [1.1891833e-18, 1.2322158e-18, 1.3274013e-18]]], dtype=float32) - bc_c1(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- bc_c1 in cloud water
- basename :
- bc_c1
array([[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 3.1663740e-28, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [2.3656756e-29, 1.4043963e-28, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - bc_c3(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- bc_c3 in cloud water
- basename :
- bc_c3
array([[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 8.7672604e-30, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [3.4671757e-30, 3.6619489e-30, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - bc_c4(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- bc_c4 in cloud water
- basename :
- bc_c4
array([[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 1.2462539e-37, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [2.0360260e-36, 7.9016866e-38, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - dei_cloud(time, lev, ncol)float3250.0 109.0 30.06 ... 27.26 27.26
- mdims :
- 9
- units :
- micrometers
- long_name :
- ice radiative effective diameter in cloud
- basename :
- dei_cloud
array([[[ 50. , 109.03206 , 30.05722 ], [ 50. , 84.69093 , 94.32067 ], [300.1505 , 95.31522 , 114.16172 ], ..., [ 27.262814, 27.262814, 27.262814], [ 27.262814, 27.262814, 27.262814], [ 27.262814, 27.262814, 27.262814]], [[ 50. , 110.49849 , 26.430277], [ 50. , 84.87456 , 110.11638 ], [167.78566 , 102.25353 , 121.98038 ], ..., [ 27.262814, 27.262814, 27.262814], [ 27.262814, 27.262814, 27.262814], [ 27.262814, 27.262814, 27.262814]], [[ 50. , 60.81266 , 23.314907], [ 50. , 92.928375, 126.68255 ], [ 50. , 44.472206, 130.24434 ], ..., ... ..., [ 27.262814, 27.262814, 27.262814], [ 27.262814, 27.262814, 27.262814], [ 27.262814, 27.262814, 27.262814]], [[ 50. , 124.17399 , 50. ], [165.49283 , 123.84373 , 50. ], [ 71.74605 , 278.94705 , 50. ], ..., [ 27.262814, 27.262814, 27.262814], [ 27.262814, 27.262814, 27.262814], [ 27.262814, 27.262814, 27.262814]], [[ 50. , 73.79445 , 50. ], [171.94188 , 44.177135, 50. ], [ 82.34086 , 36.375305, 50. ], ..., [ 27.262814, 27.262814, 27.262814], [ 27.262814, 27.262814, 27.262814], [ 27.262814, 27.262814, 27.262814]]], dtype=float32) - dgnd_a01(time, lev, ncol)float321.538e-07 1.408e-07 ... 5.35e-08
- mdims :
- 9
- units :
- m
- long_name :
- dry dgnum, interstitial, mode 01
- basename :
- dgnd_a01
array([[[1.5383517e-07, 1.4084272e-07, 1.4266240e-07], [1.5380071e-07, 1.4082329e-07, 1.4259867e-07], [1.5373723e-07, 1.4078144e-07, 1.4245236e-07], ..., [5.3499999e-08, 5.3499999e-08, 5.3499999e-08], [5.3499999e-08, 5.3499999e-08, 5.3499999e-08], [5.3499999e-08, 5.3499999e-08, 5.3499999e-08]], [[1.5361731e-07, 1.4047778e-07, 1.4292908e-07], [1.5357874e-07, 1.4045926e-07, 1.4285317e-07], [1.5350874e-07, 1.4041933e-07, 1.4267762e-07], ..., [5.3499999e-08, 5.3499999e-08, 5.3499999e-08], [5.3499999e-08, 5.3499999e-08, 5.3499999e-08], [5.3499999e-08, 5.3499999e-08, 5.3499999e-08]], [[1.5334609e-07, 1.4004537e-07, 1.4290976e-07], [1.5330185e-07, 1.4002897e-07, 1.4282678e-07], [1.5322331e-07, 1.3999347e-07, 1.4263780e-07], ..., ... ..., [5.3499999e-08, 5.3499999e-08, 5.3499999e-08], [5.3499999e-08, 5.3499999e-08, 5.3499999e-08], [5.3499999e-08, 5.3499999e-08, 5.3499999e-08]], [[1.4657347e-07, 1.4708945e-07, 1.4617697e-07], [1.4655795e-07, 1.4706691e-07, 1.4612306e-07], [1.4652153e-07, 1.4701273e-07, 1.4596088e-07], ..., [5.3499999e-08, 5.3499999e-08, 5.3499999e-08], [5.3499999e-08, 5.3499999e-08, 5.3499999e-08], [5.3499999e-08, 5.3499999e-08, 5.3499999e-08]], [[1.4628149e-07, 1.4783504e-07, 1.4563255e-07], [1.4626632e-07, 1.4781131e-07, 1.4553440e-07], [1.4623100e-07, 1.4775348e-07, 1.4524052e-07], ..., [5.3499999e-08, 5.3499999e-08, 5.3499999e-08], [5.3499999e-08, 5.3499999e-08, 5.3499999e-08], [5.3499999e-08, 5.3499999e-08, 5.3499999e-08]]], dtype=float32) - dgnd_a02(time, lev, ncol)float325.079e-08 5.064e-08 ... 1.433e-08
- mdims :
- 9
- units :
- m
- long_name :
- dry dgnum, interstitial, mode 02
- basename :
- dgnd_a02
array([[[5.07859923e-08, 5.06394287e-08, 4.92578174e-08], [5.07834379e-08, 5.06652817e-08, 4.93342078e-08], [5.07777713e-08, 5.07205726e-08, 4.95038321e-08], ..., [1.54187454e-08, 1.53957629e-08, 1.53595519e-08], [1.49048578e-08, 1.47730939e-08, 1.47801362e-08], [1.48048667e-08, 1.46658348e-08, 1.46262833e-08]], [[5.09060172e-08, 5.04418622e-08, 4.89707723e-08], [5.08978353e-08, 5.04697226e-08, 4.90758119e-08], [5.08820825e-08, 5.05293549e-08, 4.93153038e-08], ..., [1.54271991e-08, 1.53601576e-08, 1.53346615e-08], [1.50000723e-08, 1.48663482e-08, 1.48767745e-08], [1.48145034e-08, 1.47516355e-08, 1.47130832e-08]], [[5.12489784e-08, 5.00866015e-08, 4.86952771e-08], [5.12359293e-08, 5.01146076e-08, 4.88231287e-08], [5.12116607e-08, 5.01746840e-08, 4.91159327e-08], ..., ... ..., [1.50751411e-08, 1.50586406e-08, 1.50567381e-08], [1.49016000e-08, 1.48896264e-08, 1.48565800e-08], [1.45928638e-08, 1.45072141e-08, 1.44969974e-08]], [[4.12760635e-08, 5.14964782e-08, 5.10014502e-08], [4.12725392e-08, 5.14975582e-08, 5.09730285e-08], [4.12639771e-08, 5.14998817e-08, 5.08873406e-08], ..., [1.50290393e-08, 1.50410475e-08, 1.50617225e-08], [1.46867301e-08, 1.46975392e-08, 1.47830344e-08], [1.43043355e-08, 1.43159067e-08, 1.44215386e-08]], [[4.14627550e-08, 5.13728118e-08, 4.97267401e-08], [4.14612451e-08, 5.13698453e-08, 4.96583361e-08], [4.14574259e-08, 5.13617593e-08, 4.94534440e-08], ..., [1.50421808e-08, 1.50654333e-08, 1.50847388e-08], [1.45055576e-08, 1.45869654e-08, 1.47038293e-08], [1.41509542e-08, 1.42137155e-08, 1.43290038e-08]]], dtype=float32) - dgnd_a03(time, lev, ncol)float321.575e-06 1.742e-06 ... 1e-06 1e-06
- mdims :
- 9
- units :
- m
- long_name :
- dry dgnum, interstitial, mode 03
- basename :
- dgnd_a03
array([[[1.5748991e-06, 1.7423525e-06, 1.7184527e-06], [1.5739555e-06, 1.7419244e-06, 1.7183825e-06], [1.5722284e-06, 1.7409992e-06, 1.7182412e-06], ..., [1.0000000e-06, 1.0000000e-06, 1.0000000e-06], [1.0000000e-06, 1.0000000e-06, 1.0000000e-06], [1.0000000e-06, 1.0000000e-06, 1.0000000e-06]], [[1.5579387e-06, 1.7650043e-06, 1.7351489e-06], [1.5570454e-06, 1.7644320e-06, 1.7358782e-06], [1.5554376e-06, 1.7631926e-06, 1.7375719e-06], ..., [1.0000000e-06, 1.0000000e-06, 1.0000000e-06], [1.0000000e-06, 1.0000000e-06, 1.0000000e-06], [1.0000000e-06, 1.0000000e-06, 1.0000000e-06]], [[1.5515798e-06, 1.7608338e-06, 1.7442741e-06], [1.5503101e-06, 1.7603174e-06, 1.7443181e-06], [1.5480921e-06, 1.7591987e-06, 1.7442990e-06], ..., ... ..., [1.0000000e-06, 1.0000000e-06, 1.0000000e-06], [1.0000000e-06, 1.0000000e-06, 1.0000000e-06], [1.0000000e-06, 1.0000000e-06, 1.0000000e-06]], [[1.4588123e-06, 1.0838086e-06, 1.3612458e-06], [1.4582096e-06, 1.0841479e-06, 1.3619272e-06], [1.4567995e-06, 1.0849628e-06, 1.3639561e-06], ..., [1.0000000e-06, 1.0000000e-06, 1.0000000e-06], [1.0000000e-06, 1.0000000e-06, 1.0000000e-06], [1.0000000e-06, 1.0000000e-06, 1.0000000e-06]], [[1.4804053e-06, 1.0812007e-06, 1.3317351e-06], [1.4797430e-06, 1.0813245e-06, 1.3319673e-06], [1.4782057e-06, 1.0816268e-06, 1.3326531e-06], ..., [1.0000000e-06, 1.0000000e-06, 1.0000000e-06], [1.0000000e-06, 1.0000000e-06, 1.0000000e-06], [1.0000000e-06, 1.0000000e-06, 1.0000000e-06]]], dtype=float32) - dgnd_a04(time, lev, ncol)float321e-07 1e-07 ... 2.597e-08 2.619e-08
- mdims :
- 9
- units :
- m
- long_name :
- dry dgnum, interstitial, mode 04
- basename :
- dgnd_a04
array([[[1.0000000e-07, 1.0000000e-07, 1.0000000e-07], [1.0000000e-07, 1.0000000e-07, 1.0000000e-07], [1.0000000e-07, 1.0000000e-07, 1.0000000e-07], ..., [2.8425202e-08, 2.8372463e-08, 2.8288612e-08], [2.7221509e-08, 2.7023786e-08, 2.7032666e-08], [2.6974408e-08, 2.6855712e-08, 2.6811174e-08]], [[1.0000000e-07, 1.0000000e-07, 1.0000000e-07], [1.0000000e-07, 1.0000000e-07, 1.0000000e-07], [1.0000000e-07, 1.0000000e-07, 1.0000000e-07], ..., [2.8401072e-08, 2.8321045e-08, 2.8256897e-08], [2.7358036e-08, 2.7126186e-08, 2.7159155e-08], [2.6987667e-08, 2.6905669e-08, 2.6880997e-08]], [[1.0000000e-07, 1.0000000e-07, 1.0000000e-07], [1.0000000e-07, 1.0000000e-07, 1.0000000e-07], [1.0000000e-07, 1.0000000e-07, 1.0000000e-07], ..., ... ..., [2.7897389e-08, 2.7796599e-08, 2.7756624e-08], [2.7342438e-08, 2.7301324e-08, 2.7242509e-08], [2.6674346e-08, 2.6521393e-08, 2.6530563e-08]], [[1.0000000e-07, 1.0000000e-07, 1.0000000e-07], [1.0000000e-07, 1.0000000e-07, 1.0000000e-07], [1.0000000e-07, 1.0000000e-07, 1.0000000e-07], ..., [2.7869392e-08, 2.7849266e-08, 2.7855181e-08], [2.6859198e-08, 2.6901786e-08, 2.7089474e-08], [2.6147440e-08, 2.6182564e-08, 2.6377634e-08]], [[1.0000000e-07, 1.0000000e-07, 1.0000000e-07], [1.0000000e-07, 1.0000000e-07, 1.0000000e-07], [1.0000000e-07, 1.0000000e-07, 1.0000000e-07], ..., [2.7888746e-08, 2.7935044e-08, 2.7936485e-08], [2.6502741e-08, 2.6694483e-08, 2.6917320e-08], [2.5859416e-08, 2.5970316e-08, 2.6192012e-08]]], dtype=float32) - dgnw_a01(time, lev, ncol)float322.199e-07 1.797e-07 ... 5.35e-08
- mdims :
- 9
- units :
- m
- long_name :
- wet dgnum, interstitial, mode 01
- basename :
- dgnw_a01
array([[[2.1989361e-07, 1.7969839e-07, 2.0070365e-07], [2.2511547e-07, 1.8319895e-07, 2.0263751e-07], [2.3644169e-07, 1.8898365e-07, 2.0659714e-07], ..., [5.3499999e-08, 5.3499999e-08, 5.3499999e-08], [5.3499999e-08, 5.3499999e-08, 5.3499999e-08], [5.3499999e-08, 5.3499999e-08, 5.3499999e-08]], [[2.1691892e-07, 1.8235204e-07, 2.0327366e-07], [2.2172183e-07, 1.8623842e-07, 2.0538016e-07], [2.3203027e-07, 1.9235118e-07, 2.0912837e-07], ..., [5.3499999e-08, 5.3499999e-08, 5.3499999e-08], [5.3499999e-08, 5.3499999e-08, 5.3499999e-08], [5.3499999e-08, 5.3499999e-08, 5.3499999e-08]], [[2.1316407e-07, 1.8517152e-07, 2.0578611e-07], [2.1756902e-07, 1.8952119e-07, 2.0815091e-07], [2.2687054e-07, 1.9559039e-07, 2.1044234e-07], ..., ... ..., [5.3499999e-08, 5.3499999e-08, 5.3499999e-08], [5.3499999e-08, 5.3499999e-08, 5.3499999e-08], [5.3499999e-08, 5.3499999e-08, 5.3499999e-08]], [[2.3797023e-07, 2.2972235e-07, 2.2180228e-07], [2.4617412e-07, 2.3960303e-07, 2.2993616e-07], [2.6207923e-07, 2.5880956e-07, 2.4709547e-07], ..., [5.3499999e-08, 5.3499999e-08, 5.3499999e-08], [5.3499999e-08, 5.3499999e-08, 5.3499999e-08], [5.3499999e-08, 5.3499999e-08, 5.3499999e-08]], [[2.3950022e-07, 2.3801859e-07, 2.2818399e-07], [2.4795617e-07, 2.4910685e-07, 2.3833068e-07], [2.6452571e-07, 2.7143574e-07, 2.6144639e-07], ..., [5.3499999e-08, 5.3499999e-08, 5.3499999e-08], [5.3499999e-08, 5.3499999e-08, 5.3499999e-08], [5.3499999e-08, 5.3499999e-08, 5.3499999e-08]]], dtype=float32) - dgnw_a02(time, lev, ncol)float327.364e-08 6.55e-08 ... 1.433e-08
- mdims :
- 9
- units :
- m
- long_name :
- wet dgnum, interstitial, mode 02
- basename :
- dgnw_a02
array([[[7.36429087e-08, 6.55008989e-08, 7.03668022e-08], [7.53233493e-08, 6.68338132e-08, 7.11568831e-08], [7.89099133e-08, 6.90506425e-08, 7.28007308e-08], ..., [1.54187454e-08, 1.53957629e-08, 1.53595519e-08], [1.49048578e-08, 1.47730939e-08, 1.47801362e-08], [1.48048667e-08, 1.46658348e-08, 1.46262833e-08]], [[7.29825445e-08, 6.64095836e-08, 7.08074310e-08], [7.45382991e-08, 6.78798173e-08, 7.16992190e-08], [7.78274725e-08, 7.02111151e-08, 7.33841077e-08], ..., [1.54271991e-08, 1.53601576e-08, 1.53346615e-08], [1.50000723e-08, 1.48663482e-08, 1.48767745e-08], [1.48145034e-08, 1.47516355e-08, 1.47130832e-08]], [[7.23709590e-08, 6.71671216e-08, 7.13236687e-08], [7.38163521e-08, 6.87915644e-08, 7.23347426e-08], [7.68254864e-08, 7.10866885e-08, 7.36180397e-08], ..., ... ..., [1.50751411e-08, 1.50586406e-08, 1.50567381e-08], [1.49016000e-08, 1.48896264e-08, 1.48565800e-08], [1.45928638e-08, 1.45072141e-08, 1.44969974e-08]], [[6.57050165e-08, 7.98833710e-08, 7.73766615e-08], [6.77279886e-08, 8.30305140e-08, 7.99304871e-08], [7.15751369e-08, 8.90236862e-08, 8.51445563e-08], ..., [1.50290393e-08, 1.50410475e-08, 1.50617225e-08], [1.46867301e-08, 1.46975392e-08, 1.47830344e-08], [1.43043355e-08, 1.43159067e-08, 1.44215386e-08]], [[6.60981954e-08, 8.23895050e-08, 7.76356615e-08], [6.81719072e-08, 8.59018598e-08, 8.06608540e-08], [7.21597360e-08, 9.28046617e-08, 8.72290826e-08], ..., [1.50421808e-08, 1.50654333e-08, 1.50847388e-08], [1.45055576e-08, 1.45869654e-08, 1.47038293e-08], [1.41509542e-08, 1.42137155e-08, 1.43290038e-08]]], dtype=float32) - dgnw_a03(time, lev, ncol)float322.984e-06 2.777e-06 ... 1e-06 1e-06
- mdims :
- 9
- units :
- m
- long_name :
- wet dgnum, interstitial, mode 03
- basename :
- dgnw_a03
array([[[2.9843213e-06, 2.7765782e-06, 3.1700504e-06], [3.0775045e-06, 2.8557729e-06, 3.2135599e-06], [3.2767675e-06, 2.9844709e-06, 3.3028657e-06], ..., [1.0000000e-06, 1.0000000e-06, 1.0000000e-06], [1.0000000e-06, 1.0000000e-06, 1.0000000e-06], [1.0000000e-06, 1.0000000e-06, 1.0000000e-06]], [[2.9187961e-06, 2.8716329e-06, 3.2413641e-06], [3.0049403e-06, 2.9587609e-06, 3.2904056e-06], [3.1871832e-06, 3.0935041e-06, 3.3797744e-06], ..., [1.0000000e-06, 1.0000000e-06, 1.0000000e-06], [1.0000000e-06, 1.0000000e-06, 1.0000000e-06], [1.0000000e-06, 1.0000000e-06, 1.0000000e-06]], [[2.8585362e-06, 2.9339715e-06, 3.3072963e-06], [2.9380710e-06, 3.0299059e-06, 3.3608014e-06], [3.1036714e-06, 3.1617628e-06, 3.4168988e-06], ..., ... ..., [1.0000000e-06, 1.0000000e-06, 1.0000000e-06], [1.0000000e-06, 1.0000000e-06, 1.0000000e-06], [1.0000000e-06, 1.0000000e-06, 1.0000000e-06]], [[2.9261571e-06, 2.2628592e-06, 2.8831969e-06], [3.0436950e-06, 2.3850982e-06, 3.0236331e-06], [3.2698874e-06, 2.6214066e-06, 3.3209581e-06], ..., [1.0000000e-06, 1.0000000e-06, 1.0000000e-06], [1.0000000e-06, 1.0000000e-06, 1.0000000e-06], [1.0000000e-06, 1.0000000e-06, 1.0000000e-06]], [[2.9785870e-06, 2.3014518e-06, 2.9022601e-06], [3.1002694e-06, 2.4323801e-06, 3.0689666e-06], [3.3369988e-06, 2.6944581e-06, 3.4490818e-06], ..., [1.0000000e-06, 1.0000000e-06, 1.0000000e-06], [1.0000000e-06, 1.0000000e-06, 1.0000000e-06], [1.0000000e-06, 1.0000000e-06, 1.0000000e-06]]], dtype=float32) - dgnw_a04(time, lev, ncol)float321e-07 1e-07 ... 2.597e-08 2.619e-08
- mdims :
- 9
- units :
- m
- long_name :
- wet dgnum, interstitial, mode 04
- basename :
- dgnw_a04
array([[[1.00000172e-07, 1.00000044e-07, 1.00000037e-07], [1.00000186e-07, 1.00000051e-07, 1.00000044e-07], [1.00000229e-07, 1.00000058e-07, 1.00000051e-07], ..., [2.84252017e-08, 2.83724635e-08, 2.82886123e-08], [2.72215086e-08, 2.70237859e-08, 2.70326659e-08], [2.69744085e-08, 2.68557123e-08, 2.68111737e-08]], [[1.00000157e-07, 1.00000051e-07, 1.00000044e-07], [1.00000179e-07, 1.00000058e-07, 1.00000044e-07], [1.00000214e-07, 1.00000065e-07, 1.00000051e-07], ..., [2.84010717e-08, 2.83210451e-08, 2.82568973e-08], [2.73580358e-08, 2.71261857e-08, 2.71591549e-08], [2.69876672e-08, 2.69056688e-08, 2.68809970e-08]], [[1.00000150e-07, 1.00000051e-07, 1.00000044e-07], [1.00000165e-07, 1.00000058e-07, 1.00000044e-07], [1.00000193e-07, 1.00000072e-07, 1.00000051e-07], ..., ... ..., [2.78973893e-08, 2.77965988e-08, 2.77566237e-08], [2.73424376e-08, 2.73013239e-08, 2.72425087e-08], [2.66743463e-08, 2.65213931e-08, 2.65305626e-08]], [[1.00000996e-07, 1.00000229e-07, 1.00000150e-07], [1.00001124e-07, 1.00000264e-07, 1.00000179e-07], [1.00001394e-07, 1.00000349e-07, 1.00000236e-07], ..., [2.78693921e-08, 2.78492660e-08, 2.78551813e-08], [2.68591975e-08, 2.69017857e-08, 2.70894738e-08], [2.61474398e-08, 2.61825637e-08, 2.63776343e-08]], [[1.00001039e-07, 1.00000250e-07, 1.00000200e-07], [1.00001174e-07, 1.00000292e-07, 1.00000243e-07], [1.00001458e-07, 1.00000385e-07, 1.00000342e-07], ..., [2.78887455e-08, 2.79350445e-08, 2.79364851e-08], [2.65027413e-08, 2.66944831e-08, 2.69173199e-08], [2.58594159e-08, 2.59703157e-08, 2.61920121e-08]]], dtype=float32) - dst_a1(time, lev, ncol)float324.228e-12 6.471e-12 ... 2.129e-21
- mdims :
- 9
- units :
- kg/kg
- long_name :
- dst_a1 concentration
- basename :
- dst_a1
array([[[4.22774359e-12, 6.47116562e-12, 8.37841255e-12], [4.23225907e-12, 6.47699602e-12, 8.37506713e-12], [4.24103591e-12, 6.48975188e-12, 8.36269769e-12], ..., [6.44066542e-20, 5.02283681e-20, 4.66574391e-20], [8.46019893e-21, 7.88495863e-21, 7.16230651e-21], [5.80486354e-21, 7.65242879e-21, 7.80118721e-21]], [[4.22442246e-12, 5.91723068e-12, 7.93462431e-12], [4.22720929e-12, 5.92261049e-12, 7.93445865e-12], [4.23279510e-12, 5.93439837e-12, 7.92918248e-12], ..., [6.47261915e-20, 5.43362601e-20, 5.08101281e-20], [7.81291637e-21, 5.86270721e-21, 5.54297889e-21], [5.93339279e-21, 5.45236868e-21, 5.83461700e-21]], [[4.41989457e-12, 5.40385575e-12, 7.53022384e-12], [4.42209810e-12, 5.40791587e-12, 7.53286149e-12], [4.42650560e-12, 5.41684232e-12, 7.53469682e-12], ..., ... ..., [5.63053765e-20, 4.38418229e-20, 4.03745629e-20], [6.51197897e-21, 6.06556195e-21, 5.46040744e-21], [2.76640822e-21, 2.19692446e-21, 2.13495296e-21]], [[2.54932750e-12, 9.77009012e-12, 1.03578509e-11], [2.54793170e-12, 9.76853320e-12, 1.03396736e-11], [2.54472875e-12, 9.76451125e-12, 1.02846646e-11], ..., [6.08431666e-20, 5.19836195e-20, 4.92937090e-20], [4.90588624e-21, 4.68373856e-21, 5.16759116e-21], [2.30520999e-21, 2.01719988e-21, 2.11216066e-21]], [[2.21216686e-12, 8.30051999e-12, 8.65910035e-12], [2.21129582e-12, 8.29113427e-12, 8.62471119e-12], [2.20933731e-12, 8.26796010e-12, 8.52298353e-12], ..., [6.17048884e-20, 5.54316799e-20, 5.16436677e-20], [3.92348849e-21, 4.19060846e-21, 4.62160470e-21], [2.08625957e-21, 1.98178520e-21, 2.12915462e-21]]], dtype=float32) - dst_a3(time, lev, ncol)float329.09e-12 3.348e-11 ... 1.005e-36
- mdims :
- 9
- units :
- kg/kg
- long_name :
- dst_a3 concentration
- basename :
- dst_a3
array([[[9.09042459e-12, 3.34843923e-11, 4.76765259e-11], [9.11186317e-12, 3.34911925e-11, 4.79685285e-11], [9.17691357e-12, 3.35070617e-11, 4.79740171e-11], ..., [1.00000004e-36, 1.00012559e-36, 1.00008766e-36], [1.00050558e-36, 1.00000004e-36, 1.00000004e-36], [1.00000004e-36, 1.00814192e-36, 1.01708601e-36]], [[8.63755768e-12, 3.05241804e-11, 4.46189266e-11], [8.64679335e-12, 3.05179181e-11, 4.50100721e-11], [8.68594953e-12, 3.09485319e-11, 4.52389827e-11], ..., [1.00003950e-36, 1.00013797e-36, 1.00000004e-36], [1.00019653e-36, 1.00000004e-36, 1.00000004e-36], [1.00000004e-36, 1.00638619e-36, 1.02306483e-36]], [[9.81701265e-12, 2.61133389e-11, 4.16610600e-11], [9.88991094e-12, 2.72922743e-11, 4.21467063e-11], [9.87373464e-12, 2.82030545e-11, 4.22011731e-11], ..., ... ..., [1.00000004e-36, 1.00000004e-36, 1.00000004e-36], [1.00029940e-36, 1.00000004e-36, 1.00000004e-36], [1.00000004e-36, 1.00746203e-36, 1.00878010e-36]], [[9.32523590e-12, 4.28852856e-11, 5.08222527e-11], [9.35312851e-12, 4.28747246e-11, 5.07240361e-11], [9.37851879e-12, 4.28462127e-11, 5.04403255e-11], ..., [1.00003250e-36, 1.00000004e-36, 1.00000004e-36], [1.00014326e-36, 1.00006524e-36, 1.00000004e-36], [1.00229852e-36, 1.00456930e-36, 1.00836855e-36]], [[9.46585865e-12, 3.44915034e-11, 4.05322789e-11], [9.51360431e-12, 3.44441455e-11, 4.02749917e-11], [9.55412659e-12, 3.43339419e-11, 3.95399685e-11], ..., [1.00000004e-36, 1.00007663e-36, 1.00000004e-36], [1.00000004e-36, 1.00063652e-36, 1.00000004e-36], [1.00562415e-36, 1.00114215e-36, 1.00483189e-36]]], dtype=float32) - dst_c1(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- dst_c1 in cloud water
- basename :
- dst_c1
array([[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 1.2435792e-27, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [1.6499808e-28, 5.6265109e-28, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - dst_c3(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- dst_c3 in cloud water
- basename :
- dst_c3
array([[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 5.4942614e-27, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [7.2354381e-28, 2.3685013e-27, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - lambda_cloud(time, lev, ncol)float322.52e+05 0.0 0.0 ... 0.0 0.0 0.0
- mdims :
- 9
- units :
- 1/meter
- long_name :
- lambda in cloud
- basename :
- lambda_cloud
array([[[ 252000. , 0. , 0. ], [ 252000. , 0. , 0. ], [ 271525.3, 0. , 0. ], ..., [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ]], [[ 252000. , 0. , 0. ], [ 252000. , 0. , 0. ], [ 0. , 0. , 0. ], ..., [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ]], [[ 252000. , 0. , 0. ], [ 252000. , 0. , 0. ], [ 252000. , 0. , 0. ], ..., ... ..., [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ]], [[ 252000. , 0. , 252000. ], [ 0. , 0. , 252000. ], [1352728.2, 1589129.8, 252000. ], ..., [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ]], [[ 252000. , 0. , 252000. ], [ 0. , 0. , 252000. ], [1261182.2, 1534647.9, 252000. ], ..., [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ]]], dtype=float32) - mom_a1(time, lev, ncol)float326.74e-12 8.414e-12 ... 1.201e-26
- mdims :
- 9
- units :
- kg/kg
- long_name :
- mom_a1 concentration
- basename :
- mom_a1
array([[[6.73974244e-12, 8.41376621e-12, 1.04451283e-11], [6.72033653e-12, 8.40012001e-12, 1.04464016e-11], [6.68515807e-12, 8.37082140e-12, 1.04416866e-11], ..., [5.43284315e-25, 4.12282424e-25, 3.81832788e-25], [6.18996288e-26, 6.10422171e-26, 5.33022405e-26], [4.13196529e-26, 6.30235768e-26, 6.57502314e-26]], [[6.23085090e-12, 8.04079026e-12, 1.00932240e-11], [6.21409911e-12, 8.02782060e-12, 1.00987118e-11], [6.18423151e-12, 7.99989849e-12, 1.01027580e-11], ..., [5.51787003e-25, 4.50839776e-25, 4.21141529e-25], [5.51686546e-26, 4.09237495e-26, 3.75759810e-26], [4.18887698e-26, 4.16655623e-26, 4.70139622e-26]], [[5.83012891e-12, 7.57045295e-12, 9.68880705e-12], [5.81656858e-12, 7.55823789e-12, 9.69649534e-12], [5.79282802e-12, 7.53184147e-12, 9.70594612e-12], ..., ... ..., [4.75747911e-25, 3.66075981e-25, 3.37366252e-25], [4.03270070e-26, 3.77425970e-26, 3.47745541e-26], [1.61450260e-26, 1.21233315e-26, 1.16681472e-26]], [[1.19674895e-10, 7.83972470e-11, 3.61434840e-11], [1.19317875e-10, 7.83281287e-11, 3.60908074e-11], [1.18488025e-10, 7.81605683e-11, 3.59184280e-11], ..., [5.15979275e-25, 4.37857591e-25, 4.15161421e-25], [3.11353200e-26, 2.98553654e-26, 3.32795425e-26], [1.34298500e-26, 1.11663739e-26, 1.16314105e-26]], [[1.09931883e-10, 8.22941784e-11, 3.66858800e-11], [1.09591322e-10, 8.21845925e-11, 3.65697542e-11], [1.08805992e-10, 8.19156687e-11, 3.62231044e-11], ..., [5.23523398e-25, 4.65503073e-25, 4.33683925e-25], [2.44170045e-26, 2.64304981e-26, 2.97512635e-26], [1.24100609e-26, 1.12704927e-26, 1.20109512e-26]]], dtype=float32) - mom_a2(time, lev, ncol)float323.87e-14 4.78e-14 ... 8.16e-21
- mdims :
- 9
- units :
- kg/kg
- long_name :
- mom_a2 concentration
- basename :
- mom_a2
array([[[3.86956905e-14, 4.77997768e-14, 5.76490827e-14], [3.82888166e-14, 4.75248435e-14, 5.78104865e-14], [3.75505596e-14, 4.69597505e-14, 5.81481410e-14], ..., [1.30988808e-20, 1.28381977e-20, 1.25610510e-20], [1.04955883e-20, 9.94421992e-21, 1.00050266e-20], [1.01006201e-20, 9.71198896e-21, 9.67330615e-21]], [[3.07236909e-14, 4.17096506e-14, 5.17551496e-14], [3.03936631e-14, 4.14521797e-14, 5.19410056e-14], [2.98049617e-14, 4.09295604e-14, 5.23413981e-14], ..., [1.30307079e-20, 1.26212696e-20, 1.24456092e-20], [1.09365467e-20, 1.04031169e-20, 1.05054668e-20], [1.01552180e-20, 1.01735477e-20, 1.01930543e-20]], [[2.33709536e-14, 3.57082561e-14, 4.57672637e-14], [2.31210975e-14, 3.54796724e-14, 4.59494740e-14], [2.26865510e-14, 3.50224847e-14, 4.63734716e-14], ..., ... ..., [1.08579556e-20, 1.06584193e-20, 1.06412480e-20], [1.03132774e-20, 1.03258781e-20, 1.02129300e-20], [9.20473823e-21, 9.07176086e-21, 9.06547381e-21]], [[7.78547311e-13, 4.04072229e-13, 1.75003729e-13], [7.72750353e-13, 4.02636393e-13, 1.74836016e-13], [7.59937035e-13, 3.99391918e-13, 1.74329531e-13], ..., [1.08176354e-20, 1.08366347e-20, 1.08946626e-20], [9.44316738e-21, 9.51534535e-21, 9.86122236e-21], [8.00108300e-21, 8.14807558e-21, 8.67260825e-21]], [[6.78530962e-13, 3.60345976e-13, 1.35161833e-13], [6.73195766e-13, 3.58792369e-13, 1.34174423e-13], [6.61450332e-13, 3.55248491e-13, 1.31293834e-13], ..., [1.08337929e-20, 1.10221057e-20, 1.11158203e-20], [8.59199126e-21, 8.97693559e-21, 9.51978499e-21], [7.33993870e-21, 7.59276274e-21, 8.16048086e-21]]], dtype=float32) - mom_a3(time, lev, ncol)float322.536e-13 1.29e-12 ... 1.005e-36
- mdims :
- 9
- units :
- kg/kg
- long_name :
- mom_a3 concentration
- basename :
- mom_a3
array([[[2.53627441e-13, 1.29007536e-12, 1.94297855e-12], [2.58551806e-13, 1.28624856e-12, 1.97905234e-12], [2.79789267e-13, 1.27802304e-12, 2.00091788e-12], ..., [1.00000004e-36, 1.00012559e-36, 1.00008766e-36], [1.00050558e-36, 1.00000004e-36, 1.00000004e-36], [1.00000004e-36, 1.00814192e-36, 1.01708601e-36]], [[2.47831703e-13, 1.53423433e-12, 2.11366406e-12], [2.51278165e-13, 1.52796981e-12, 2.16676265e-12], [2.67773867e-13, 1.60550468e-12, 2.22235381e-12], ..., [1.00003950e-36, 1.00013797e-36, 1.00000004e-36], [1.00019653e-36, 1.00000004e-36, 1.00000004e-36], [1.00000004e-36, 1.00638619e-36, 1.02306483e-36]], [[5.84717875e-13, 1.43747601e-12, 2.23248351e-12], [6.20913368e-13, 1.69541952e-12, 2.29164821e-12], [6.19221905e-13, 1.88974270e-12, 2.31600980e-12], ..., ... ..., [1.00000004e-36, 1.00000004e-36, 1.00000004e-36], [1.00029940e-36, 1.00000004e-36, 1.00000004e-36], [1.00000004e-36, 1.00746203e-36, 1.00878010e-36]], [[6.29818680e-13, 5.95921912e-13, 6.08017217e-13], [6.34925435e-13, 6.00955863e-13, 6.05474492e-13], [6.31018675e-13, 6.07442861e-13, 5.98078661e-13], ..., [1.00003250e-36, 1.00000004e-36, 1.00000004e-36], [1.00014326e-36, 1.00006524e-36, 1.00000004e-36], [1.00229852e-36, 1.00456930e-36, 1.00836855e-36]], [[6.75472102e-13, 5.45208030e-13, 5.13321211e-13], [6.82480927e-13, 5.45398199e-13, 5.08033773e-13], [6.79733125e-13, 5.42964545e-13, 4.94894436e-13], ..., [1.00000004e-36, 1.00007663e-36, 1.00000004e-36], [1.00000004e-36, 1.00063652e-36, 1.00000004e-36], [1.00562415e-36, 1.00114215e-36, 1.00483189e-36]]], dtype=float32) - mom_a4(time, lev, ncol)float325.877e-17 6.053e-17 ... 3.232e-23
- mdims :
- 9
- units :
- kg/kg
- long_name :
- mom_a4 concentration
- basename :
- mom_a4
array([[[5.87696671e-17, 6.05256525e-17, 4.84329800e-17], [5.88308322e-17, 6.05343610e-17, 4.84862239e-17], [5.89544394e-17, 6.05611948e-17, 4.85605411e-17], ..., [5.42586293e-23, 5.32887469e-23, 5.20619104e-23], [4.19554536e-23, 3.94910048e-23, 3.97038711e-23], [4.00615280e-23, 3.84254778e-23, 3.82540299e-23]], [[5.91907021e-17, 6.39834792e-17, 5.24451070e-17], [5.92515759e-17, 6.39971177e-17, 5.25274148e-17], [5.93707958e-17, 6.40324416e-17, 5.26526764e-17], ..., [5.39316496e-23, 5.23187036e-23, 5.15142075e-23], [4.38300364e-23, 4.13142423e-23, 4.17341308e-23], [4.03459344e-23, 4.01616092e-23, 4.02136929e-23]], [[5.91552987e-17, 6.67982094e-17, 5.60775119e-17], [5.92185416e-17, 6.68163081e-17, 5.61824315e-17], [5.93376160e-17, 6.68604398e-17, 5.63247992e-17], ..., ... ..., [4.55809227e-23, 4.46206486e-23, 4.44418328e-23], [4.19759356e-23, 4.18209560e-23, 4.12792610e-23], [3.65126195e-23, 3.57559977e-23, 3.58116219e-23]], [[1.61680511e-16, 1.31672039e-16, 1.15966500e-16], [1.61689219e-16, 1.31709031e-16, 1.16128151e-16], [1.61748856e-16, 1.31825524e-16, 1.16619562e-16], ..., [4.52957463e-23, 4.53099742e-23, 4.55247937e-23], [3.78179571e-23, 3.81140955e-23, 3.97217719e-23], [3.16518321e-23, 3.21944832e-23, 3.42584905e-23]], [[1.92864280e-16, 1.30462251e-16, 1.21383448e-16], [1.92892973e-16, 1.30546663e-16, 1.21569914e-16], [1.93007085e-16, 1.30662720e-16, 1.22129234e-16], ..., [4.53953416e-23, 4.61577599e-23, 4.65010500e-23], [3.43152443e-23, 3.59766674e-23, 3.82451284e-23], [2.90534979e-23, 3.00771585e-23, 3.23208398e-23]]], dtype=float32) - mom_c1(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- mom_c1 in cloud water
- basename :
- mom_c1
array([[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 9.8904715e-27, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [8.0522761e-27, 5.5510986e-27, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - mom_c2(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- mom_c2 in cloud water
- basename :
- mom_c2
array([[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 4.5407404e-29, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [3.9663239e-29, 2.1883019e-29, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - mom_c3(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- mom_c3 in cloud water
- basename :
- mom_c3
array([[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 7.1733096e-29, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [4.7518111e-29, 3.5826188e-29, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - mom_c4(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- mom_c4 in cloud water
- basename :
- mom_c4
array([[[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., ... ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]]], dtype=float32) - mu_cloud(time, lev, ncol)float325.3 0.0 0.0 5.3 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- 1
- long_name :
- mu in cloud
- basename :
- mu_cloud
array([[[ 5.3 , 0. , 0. ], [ 5.3 , 0. , 0. ], [12.576265, 0. , 0. ], ..., [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ]], [[ 5.3 , 0. , 0. ], [ 5.3 , 0. , 0. ], [ 0. , 0. , 0. ], ..., [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ]], [[ 5.3 , 0. , 0. ], [ 5.3 , 0. , 0. ], [ 5.3 , 0. , 0. ], ..., ... ..., [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ]], [[ 5.3 , 0. , 5.3 ], [ 0. , 0. , 5.3 ], [12.576265, 12.576265, 5.3 ], ..., [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ]], [[ 5.3 , 0. , 5.3 ], [ 0. , 0. , 5.3 ], [12.576265, 12.576265, 5.3 ], ..., [ 0. , 0. , 0. ], [ 0. , 0. , 0. ], [ 0. , 0. , 0. ]]], dtype=float32) - ncl_a1(time, lev, ncol)float324.32e-11 5.234e-11 ... 1.155e-21
- mdims :
- 9
- units :
- kg/kg
- long_name :
- ncl_a1 concentration
- basename :
- ncl_a1
array([[[4.32010261e-11, 5.23371763e-11, 5.88009294e-11], [4.31280359e-11, 5.22596653e-11, 5.86116503e-11], [4.29956140e-11, 5.20936037e-11, 5.81548316e-11], ..., [5.40016785e-20, 4.15065531e-20, 3.83975172e-20], [5.69262224e-21, 5.54135562e-21, 4.85878339e-21], [3.70428886e-21, 5.54755625e-21, 5.73263306e-21]], [[4.05375213e-11, 4.99237075e-11, 5.64955756e-11], [4.04708454e-11, 4.98466650e-11, 5.63064352e-11], [4.03516838e-11, 4.96812522e-11, 5.58476078e-11], ..., [5.44273275e-20, 4.54322134e-20, 4.24614362e-20], [5.02175695e-21, 3.61854481e-21, 3.31904848e-21], [3.84360579e-21, 3.61779719e-21, 4.04152046e-21]], [[3.86611368e-11, 4.71960838e-11, 5.39920192e-11], [3.86041824e-11, 4.71190344e-11, 5.38170411e-11], [3.85039431e-11, 4.69530803e-11, 5.33996736e-11], ..., ... ..., [4.73564810e-20, 3.64721930e-20, 3.36802807e-20], [3.82582060e-21, 3.62043706e-21, 3.38505107e-21], [1.51689001e-21, 1.14966483e-21, 1.09719823e-21]], [[1.55931351e-10, 1.05489527e-10, 5.77640817e-11], [1.55340130e-10, 1.05219770e-10, 5.74666773e-11], [1.53968005e-10, 1.04573468e-10, 5.65823638e-11], ..., [5.17737903e-20, 4.40417938e-20, 4.17247640e-20], [3.05171970e-21, 2.95922370e-21, 3.26860419e-21], [1.30500010e-21, 1.07686506e-21, 1.10578205e-21]], [[1.49276910e-10, 1.12530790e-10, 5.44484215e-11], [1.48692628e-10, 1.12199389e-10, 5.40401265e-11], [1.47347592e-10, 1.11398675e-10, 5.28444510e-11], ..., [5.28216860e-20, 4.70737694e-20, 4.37682943e-20], [2.41742403e-21, 2.62287966e-21, 2.90901852e-21], [1.22038241e-21, 1.09821030e-21, 1.15463629e-21]]], dtype=float32) - ncl_a2(time, lev, ncol)float321.85e-13 2.948e-13 ... 4.3e-16
- mdims :
- 9
- units :
- kg/kg
- long_name :
- ncl_a2 concentration
- basename :
- ncl_a2
array([[[1.8502070e-13, 2.9483504e-13, 3.5232109e-13], [1.8351752e-13, 2.9312541e-13, 3.5007359e-13], [1.8078198e-13, 2.8961485e-13, 3.4503766e-13], ..., [6.5833831e-16, 6.4291442e-16, 6.3044223e-16], [5.4672710e-16, 5.1803355e-16, 5.2097391e-16], [5.2951402e-16, 5.0561535e-16, 5.0292819e-16]], [[1.4996630e-13, 2.6230670e-13, 3.2053028e-13], [1.4877214e-13, 2.6064215e-13, 3.1801037e-13], [1.4663298e-13, 2.5726402e-13, 3.1240112e-13], ..., [6.5689115e-16, 6.3332452e-16, 6.2588519e-16], [5.6943574e-16, 5.4348704e-16, 5.4854155e-16], [5.3167252e-16, 5.3292354e-16, 5.3263036e-16]], [[1.2019540e-13, 2.3291969e-13, 2.8831045e-13], [1.1931058e-13, 2.3135763e-13, 2.8576927e-13], [1.1776185e-13, 2.2822998e-13, 2.8040520e-13], ..., ... ..., [5.4301398e-16, 5.3801214e-16, 5.3908035e-16], [5.2943964e-16, 5.3167485e-16, 5.2711369e-16], [4.8091556e-16, 4.7606862e-16, 4.7432347e-16]], [[1.3732685e-12, 6.7783019e-13, 3.2273208e-13], [1.3618829e-12, 6.7380926e-13, 3.2028490e-13], [1.3366305e-12, 6.6454763e-13, 3.1323872e-13], ..., [5.3974835e-16, 5.4333685e-16, 5.4700927e-16], [4.9291669e-16, 4.9580110e-16, 5.1059199e-16], [4.2142499e-16, 4.2881805e-16, 4.5534019e-16]], [[1.2886010e-12, 6.7415935e-13, 2.7516281e-13], [1.2774375e-12, 6.6960592e-13, 2.7167073e-13], [1.2527985e-12, 6.5908422e-13, 2.6163094e-13], ..., [5.3638859e-16, 5.4678200e-16, 5.5367013e-16], [4.5119868e-16, 4.6918590e-16, 4.9528626e-16], [3.8744857e-16, 4.0114737e-16, 4.3004882e-16]]], dtype=float32) - ncl_a3(time, lev, ncol)float328.369e-10 1.43e-09 ... 1.005e-36
- mdims :
- 9
- units :
- kg/kg
- long_name :
- ncl_a3 concentration
- basename :
- ncl_a3
array([[[8.36920033e-10, 1.43039613e-09, 1.59091040e-09], [8.35040925e-10, 1.42707701e-09, 1.58781810e-09], [8.33173364e-10, 1.41997925e-09, 1.57293834e-09], ..., [1.00000004e-36, 1.00012559e-36, 1.00008766e-36], [1.00050558e-36, 1.00000004e-36, 1.00000004e-36], [1.00000004e-36, 1.00814192e-36, 1.01708601e-36]], [[7.54990848e-10, 1.40291578e-09, 1.56323476e-09], [7.53272278e-10, 1.39927991e-09, 1.56220958e-09], [7.51473939e-10, 1.40513445e-09, 1.55130808e-09], ..., [1.00003950e-36, 1.00013797e-36, 1.00000004e-36], [1.00019653e-36, 1.00000004e-36, 1.00000004e-36], [1.00000004e-36, 1.00638619e-36, 1.02306483e-36]], [[7.26354754e-10, 1.31276168e-09, 1.50954671e-09], [7.28928584e-10, 1.34872891e-09, 1.50913204e-09], [7.25543237e-10, 1.37198330e-09, 1.49425905e-09], ..., ... ..., [1.00000004e-36, 1.00000004e-36, 1.00000004e-36], [1.00029940e-36, 1.00000004e-36, 1.00000004e-36], [1.00000004e-36, 1.00746203e-36, 1.00878010e-36]], [[2.55126564e-09, 7.17840842e-10, 7.76307130e-10], [2.53750909e-09, 7.16333937e-10, 7.73043130e-10], [2.50605958e-09, 7.12863435e-10, 7.63481667e-10], ..., [1.00003250e-36, 1.00000004e-36, 1.00000004e-36], [1.00014326e-36, 1.00006524e-36, 1.00000004e-36], [1.00229852e-36, 1.00456930e-36, 1.00836855e-36]], [[2.52600918e-09, 7.61576746e-10, 6.78960832e-10], [2.51197774e-09, 7.59146357e-10, 6.73573641e-10], [2.48005239e-09, 7.53566209e-10, 6.58102628e-10], ..., [1.00000004e-36, 1.00007663e-36, 1.00000004e-36], [1.00000004e-36, 1.00063652e-36, 1.00000004e-36], [1.00562415e-36, 1.00114215e-36, 1.00483189e-36]]], dtype=float32) - ncl_c1(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- ncl_c1 in cloud water
- basename :
- ncl_c1
array([[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 1.3073943e-26, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [1.0845050e-26, 7.4780355e-27, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - ncl_c2(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- ncl_c2 in cloud water
- basename :
- ncl_c2
array([[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 7.4214928e-29, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [7.4652975e-29, 3.9946077e-29, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - ncl_c3(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- ncl_c3 in cloud water
- basename :
- ncl_c3
array([[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 9.0498302e-26, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [1.8302684e-25, 5.1297905e-26, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - num_a1(time, lev, ncol)float321.86e+07 2.059e+07 ... 0.04611
- mdims :
- 9
- units :
- 1/kg
- long_name :
- num_a1 concentration
- basename :
- num_a1
array([[[1.8599182e+07, 2.0588332e+07, 2.3138102e+07], [1.8613988e+07, 2.0603666e+07, 2.3147224e+07], [1.8642532e+07, 2.0634804e+07, 2.3152100e+07], ..., [1.4345158e+00, 1.1117719e+00, 1.0307112e+00], [1.9578731e-01, 1.6762729e-01, 1.4971410e-01], [1.3382529e-01, 1.7172536e-01, 1.7721485e-01]], [[1.8742460e+07, 1.9788916e+07, 2.2417596e+07], [1.8753600e+07, 1.9804890e+07, 2.2439056e+07], [1.8775244e+07, 1.9836370e+07, 2.2470252e+07], ..., [1.4458982e+00, 1.2093395e+00, 1.1307710e+00], [1.8763921e-01, 1.3313664e-01, 1.2553309e-01], [1.4174139e-01, 1.2061103e-01, 1.2853038e-01]], [[1.9397822e+07, 1.9002198e+07, 2.1775338e+07], [1.9408690e+07, 1.9016370e+07, 2.1806410e+07], [1.9429084e+07, 1.9042894e+07, 2.1855532e+07], ..., ... ..., [1.2519724e+00, 9.7528940e-01, 9.0460289e-01], [1.5025558e-01, 1.4195167e-01, 1.3279216e-01], [5.8234263e-02, 4.5864798e-02, 4.4114098e-02]], [[3.0289586e+07, 3.5580064e+07, 3.0782620e+07], [3.0230462e+07, 3.5577348e+07, 3.0738350e+07], [3.0089832e+07, 3.5567864e+07, 3.0600744e+07], ..., [1.3777106e+00, 1.1806866e+00, 1.1186939e+00], [1.2906876e-01, 1.2248679e-01, 1.2943783e-01], [5.0766341e-02, 4.3339692e-02, 4.4507414e-02]], [[2.8468060e+07, 3.4611704e+07, 2.8127992e+07], [2.8409266e+07, 3.4586636e+07, 2.8050288e+07], [2.8270700e+07, 3.4521740e+07, 2.7818734e+07], ..., [1.4122443e+00, 1.2664983e+00, 1.1764649e+00], [1.1244163e-01, 1.1266320e-01, 1.1681487e-01], [4.7260772e-02, 4.3706317e-02, 4.6113059e-02]]], dtype=float32) - num_a2(time, lev, ncol)float321.002e+07 1.586e+07 ... 3.83e+06
- mdims :
- 9
- units :
- 1/kg
- long_name :
- num_a2 concentration
- basename :
- num_a2
array([[[10024589. , 15859493. , 22432274. ], [10027550. , 15829427. , 22187108. ], [10034107. , 15767270. , 21641080. ], ..., [ 4753257. , 4667846. , 4612473. ], [ 4350748.5, 4200435.5, 4219150.5], [ 4295866. , 4169037.2, 4173743. ]], [[10026496. , 15626587. , 22667688. ], [10030834. , 15593760. , 22343884. ], [10039808. , 15526264. , 21618768. ], ..., [ 4740030.5, 4629462.5, 4599416. ], [ 4461686. , 4345496. , 4376109. ], [ 4314747.5, 4340131. , 4358134. ]], [[10175696. , 15619343. , 22928344. ], [10181941. , 15583175. , 22533300. ], [10194128. , 15509105. , 21661414. ], ..., ... ..., [ 4197740.5, 4172162.2, 4179986. ], [ 4198904. , 4217172.5, 4206137. ], [ 4020058. , 4041889.5, 4039863.5]], [[32994956. , 22667226. , 22555044. ], [32919884. , 22649126. , 22522964. ], [32748878. , 22607664. , 22427530. ], ..., [ 4207644.5, 4221433. , 4229333.5], [ 4077168.5, 4086484.8, 4134859.5], [ 3774417.2, 3823092.2, 3953772. ]], [[31712162. , 22356636. , 20903662. ], [31634676. , 22328604. , 20844644. ], [31459214. , 22263176. , 20672252. ], ..., [ 4163785.8, 4217300. , 4250392. ], [ 3888963.5, 3971150.2, 4083001.2], [ 3605583.5, 3677227.5, 3829992.8]]], dtype=float32) - num_a3(time, lev, ncol)float324.635e+04 5.995e+04 ... 1e-05 1e-05
- mdims :
- 9
- units :
- 1/kg
- long_name :
- num_a3 concentration
- basename :
- num_a3
array([[[4.6353160e+04, 5.9953895e+04, 6.9670859e+04], [4.6287258e+04, 5.9860637e+04, 6.9385852e+04], [4.6167824e+04, 5.9661645e+04, 6.8708906e+04], ..., [9.9999997e-06, 9.9999997e-06, 9.9999997e-06], [9.9999997e-06, 9.9999997e-06, 9.9999997e-06], [9.9999997e-06, 9.9999997e-06, 9.9999997e-06]], [[4.3343113e+04, 5.6853492e+04, 6.6656906e+04], [4.3283758e+04, 5.6760871e+04, 6.6364242e+04], [4.3178164e+04, 5.6561922e+04, 6.5666906e+04], ..., [9.9999997e-06, 9.9999997e-06, 9.9999997e-06], [9.9999997e-06, 9.9999997e-06, 9.9999997e-06], [9.9999997e-06, 9.9999997e-06, 9.9999997e-06]], [[4.1218855e+04, 5.3480301e+04, 6.3404270e+04], [4.1168973e+04, 5.3388109e+04, 6.3123668e+04], [4.1082191e+04, 5.3189648e+04, 6.2466926e+04], ..., ... ..., [9.9999997e-06, 9.9999997e-06, 9.9999997e-06], [9.9999997e-06, 9.9999997e-06, 9.9999997e-06], [9.9999997e-06, 9.9999997e-06, 9.9999997e-06]], [[1.7676647e+05, 1.2761860e+05, 6.9958312e+04], [1.7602891e+05, 1.2724487e+05, 6.9566820e+04], [1.7435706e+05, 1.2636881e+05, 6.8419844e+04], ..., [9.9999997e-06, 9.9999997e-06, 9.9999997e-06], [9.9999997e-06, 9.9999997e-06, 9.9999997e-06], [9.9999997e-06, 9.9999997e-06, 9.9999997e-06]], [[1.6736406e+05, 1.3479756e+05, 6.5092113e+04], [1.6665097e+05, 1.3434503e+05, 6.4546461e+04], [1.6504758e+05, 1.3328209e+05, 6.2973477e+04], ..., [9.9999997e-06, 9.9999997e-06, 9.9999997e-06], [9.9999997e-06, 9.9999997e-06, 9.9999997e-06], [9.9999997e-06, 9.9999997e-06, 9.9999997e-06]]], dtype=float32) - num_a4(time, lev, ncol)float322.31e+06 4.839e+06 ... 1.078e+03
- mdims :
- 9
- units :
- 1/kg
- long_name :
- num_a4 concentration
- basename :
- num_a4
array([[[2.31008075e+06, 4.83856050e+06, 7.67385550e+06], [2.31257050e+06, 4.82402600e+06, 7.51563600e+06], [2.31763600e+06, 4.79391650e+06, 7.16570150e+06], ..., [1.41482361e+03, 1.39863037e+03, 1.37913684e+03], [1.24484656e+03, 1.19543079e+03, 1.20032971e+03], [1.22054553e+03, 1.18377454e+03, 1.18423486e+03]], [[2.40009775e+06, 5.20991000e+06, 8.36777500e+06], [2.40235425e+06, 5.19423550e+06, 8.16266650e+06], [2.40688375e+06, 5.16213750e+06, 7.70587550e+06], ..., [1.41024927e+03, 1.38069312e+03, 1.36920020e+03], [1.28139185e+03, 1.23698022e+03, 1.24495813e+03], [1.22868750e+03, 1.23015857e+03, 1.23426038e+03]], [[2.47771725e+06, 5.58883600e+06, 9.00453100e+06], [2.47969625e+06, 5.57210250e+06, 8.75786000e+06], [2.48361525e+06, 5.53810900e+06, 8.21539400e+06], ..., ... ..., [1.26301575e+03, 1.25043176e+03, 1.25114075e+03], [1.23028699e+03, 1.22948291e+03, 1.22136023e+03], [1.14848157e+03, 1.14360742e+03, 1.14460217e+03]], [[1.05484400e+06, 4.34455600e+06, 6.60874650e+06], [1.05484900e+06, 4.33365200e+06, 6.56882700e+06], [1.05508075e+06, 4.30765200e+06, 6.45045700e+06], ..., [1.25766992e+03, 1.26153186e+03, 1.26646301e+03], [1.16843701e+03, 1.17191968e+03, 1.19643896e+03], [1.06112927e+03, 1.07397681e+03, 1.11542114e+03]], [[1.28474888e+06, 4.09214650e+06, 5.59878200e+06], [1.28506375e+06, 4.08161750e+06, 5.54798100e+06], [1.28607012e+06, 4.05651025e+06, 5.39911500e+06], ..., [1.25558167e+03, 1.27072522e+03, 1.28041357e+03], [1.10636780e+03, 1.13499927e+03, 1.17489294e+03], [1.00912646e+03, 1.03072681e+03, 1.07770020e+03]]], dtype=float32) - num_c1(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- 1/kg
- long_name :
- num_c1 in cloud water
- basename :
- num_c1
array([[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 4.3198689e-09, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [2.0086866e-09, 2.2560782e-09, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - num_c2(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- 1/kg
- long_name :
- num_c2 in cloud water
- basename :
- num_c2
array([[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 1.3501112e-09, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [7.0704365e-10, 7.4747025e-10, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - num_c3(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- 1/kg
- long_name :
- num_c3 in cloud water
- basename :
- num_c3
array([[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 1.5845851e-11, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [1.2213930e-11, 8.9876700e-12, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - num_c4(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- 1/kg
- long_name :
- num_c4 in cloud water
- basename :
- num_c4
array([[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 4.3307552e-23, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [4.7332813e-21, 2.8289060e-23, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - pom_a1(time, lev, ncol)float322.438e-12 4.334e-12 ... 2.49e-21
- mdims :
- 9
- units :
- kg/kg
- long_name :
- pom_a1 concentration
- basename :
- pom_a1
array([[[2.4381541e-12, 4.3339893e-12, 5.5313588e-12], [2.4450580e-12, 4.3386297e-12, 5.5274856e-12], [2.4577538e-12, 4.3479985e-12, 5.5139170e-12], ..., [8.3421236e-20, 6.5956248e-20, 6.1498940e-20], [9.8092804e-21, 8.9135165e-21, 8.0440261e-21], [6.4215077e-21, 8.5472371e-21, 8.7378796e-21]], [[2.4367431e-12, 3.9552901e-12, 5.1916093e-12], [2.4434075e-12, 3.9600719e-12, 5.1897306e-12], [2.4555690e-12, 3.9693392e-12, 5.1800040e-12], ..., [8.3330556e-20, 7.1535342e-20, 6.7004544e-20], [8.8798881e-21, 6.4097333e-21, 6.0145979e-21], [6.7386551e-21, 5.8814997e-21, 6.2966018e-21]], [[2.5618763e-12, 3.6004164e-12, 4.8799094e-12], [2.5686161e-12, 3.6047456e-12, 4.8796774e-12], [2.5806579e-12, 3.6126857e-12, 4.8724835e-12], ..., ... ..., [7.2520230e-20, 5.6722734e-20, 5.2587015e-20], [7.3997881e-21, 6.9874014e-21, 6.4938767e-21], [3.1362359e-21, 2.5036188e-21, 2.3959945e-21]], [[1.8689733e-12, 7.4346362e-12, 8.2421275e-12], [1.8681710e-12, 7.4330707e-12, 8.2249598e-12], [1.8660475e-12, 7.4289628e-12, 8.1727793e-12], ..., [7.9183822e-20, 6.8224526e-20, 6.4815340e-20], [5.9082656e-21, 5.7381341e-21, 6.2607152e-21], [2.7226418e-21, 2.3492951e-21, 2.4175725e-21]], [[1.5934742e-12, 6.3371166e-12, 6.8903052e-12], [1.5931622e-12, 6.3298308e-12, 6.8589752e-12], [1.5921261e-12, 6.3115255e-12, 6.7659645e-12], ..., [8.0518032e-20, 7.2850378e-20, 6.8054947e-20], [4.7836618e-21, 5.1409675e-21, 5.5862172e-21], [2.4981919e-21, 2.3450758e-21, 2.4899839e-21]]], dtype=float32) - pom_a3(time, lev, ncol)float321.1e-13 7.726e-13 ... 1.005e-36
- mdims :
- 9
- units :
- kg/kg
- long_name :
- pom_a3 concentration
- basename :
- pom_a3
array([[[1.10009698e-13, 7.72626050e-13, 1.17652719e-12], [1.11981130e-13, 7.70656163e-13, 1.21709500e-12], [1.20121429e-13, 7.66420944e-13, 1.23776542e-12], ..., [1.00000004e-36, 1.00012559e-36, 1.00008766e-36], [1.00050558e-36, 1.00000004e-36, 1.00000004e-36], [1.00000004e-36, 1.00814192e-36, 1.01708601e-36]], [[1.02022103e-13, 9.00941973e-13, 1.26257175e-12], [1.03590632e-13, 8.97539421e-13, 1.31423117e-12], [1.10508845e-13, 9.46212835e-13, 1.35817063e-12], ..., [1.00003950e-36, 1.00013797e-36, 1.00000004e-36], [1.00019653e-36, 1.00000004e-36, 1.00000004e-36], [1.00000004e-36, 1.00638619e-36, 1.02306483e-36]], [[2.43686283e-13, 8.09313933e-13, 1.33039044e-12], [2.60037760e-13, 9.64827068e-13, 1.39287225e-12], [2.59908685e-13, 1.08272159e-12, 1.41164034e-12], ..., ... ..., [1.00000004e-36, 1.00000004e-36, 1.00000004e-36], [1.00029940e-36, 1.00000004e-36, 1.00000004e-36], [1.00000004e-36, 1.00746203e-36, 1.00878010e-36]], [[1.23594711e-13, 2.32022518e-13, 2.76576179e-13], [1.24698917e-13, 2.32865132e-13, 2.75625307e-13], [1.23885047e-13, 2.33870188e-13, 2.72870539e-13], ..., [1.00003250e-36, 1.00000004e-36, 1.00000004e-36], [1.00014326e-36, 1.00006524e-36, 1.00000004e-36], [1.00229852e-36, 1.00456930e-36, 1.00836855e-36]], [[1.56974517e-13, 1.85233922e-13, 2.12604443e-13], [1.58968772e-13, 1.85145044e-13, 2.10560898e-13], [1.58276766e-13, 1.84341108e-13, 2.05185614e-13], ..., [1.00000004e-36, 1.00007663e-36, 1.00000004e-36], [1.00000004e-36, 1.00063652e-36, 1.00000004e-36], [1.00562415e-36, 1.00114215e-36, 1.00483189e-36]]], dtype=float32) - pom_a4(time, lev, ncol)float322.182e-12 4.21e-12 ... 2.662e-17
- mdims :
- 9
- units :
- kg/kg
- long_name :
- pom_a4 concentration
- basename :
- pom_a4
array([[[2.1824196e-12, 4.2101236e-12, 6.4961925e-12], [2.1846917e-12, 4.1980490e-12, 6.3637316e-12], [2.1893305e-12, 4.1730738e-12, 6.0707073e-12], ..., [4.4655438e-17, 4.3900950e-17, 4.2907844e-17], [3.4512870e-17, 3.2422128e-17, 3.2586615e-17], [3.2925279e-17, 3.1508143e-17, 3.1363284e-17]], [[2.2761643e-12, 4.5733009e-12, 7.0652052e-12], [2.2782414e-12, 4.5603699e-12, 6.8951607e-12], [2.2824212e-12, 4.5339657e-12, 6.5163716e-12], ..., [4.4399184e-17, 4.3102830e-17, 4.2455478e-17], [3.6064066e-17, 3.3933586e-17, 3.4277034e-17], [3.3196065e-17, 3.2926632e-17, 3.2943722e-17]], [[2.3483682e-12, 4.9426041e-12, 7.5956593e-12], [2.3502439e-12, 4.9289301e-12, 7.3924729e-12], [2.3539530e-12, 4.9012738e-12, 6.9455847e-12], ..., ... ..., [3.7696348e-17, 3.6920370e-17, 3.6783058e-17], [3.4566127e-17, 3.4384714e-17, 3.3936967e-17], [2.9954354e-17, 2.9316280e-17, 2.9372859e-17]], [[1.5040958e-12, 4.5600832e-12, 6.2887560e-12], [1.5039347e-12, 4.5481774e-12, 6.2496995e-12], [1.5038630e-12, 4.5197917e-12, 6.1341427e-12], ..., [3.7419050e-17, 3.7455770e-17, 3.7626213e-17], [3.1117426e-17, 3.1359046e-17, 3.2690373e-17], [2.6075211e-17, 2.6495808e-17, 2.8134090e-17]], [[1.7574820e-12, 4.4061066e-12, 5.4620011e-12], [1.7576609e-12, 4.3944440e-12, 5.4132775e-12], [1.7584460e-12, 4.3666550e-12, 5.2706641e-12], ..., [3.7428080e-17, 3.8070829e-17, 3.8369196e-17], [2.8312460e-17, 2.9680084e-17, 3.1495296e-17], [2.3990259e-17, 2.4819273e-17, 2.6617248e-17]]], dtype=float32) - pom_c1(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- pom_c1 in cloud water
- basename :
- pom_c1
array([[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 9.4409847e-28, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [1.1925509e-28, 4.2944116e-28, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - pom_c3(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- pom_c3 in cloud water
- basename :
- pom_c3
array([[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 2.8861583e-29, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [1.0847047e-29, 1.2401963e-29, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - pom_c4(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- pom_c4 in cloud water
- basename :
- pom_c4
array([[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 1.1214540e-37, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [5.6138077e-36, 7.2188423e-38, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - so4_a1(time, lev, ncol)float329.182e-11 5.125e-11 ... 5.673e-21
- mdims :
- 9
- units :
- kg/kg
- long_name :
- so4_a1 concentration
- basename :
- so4_a1
array([[[9.18248255e-11, 5.12464585e-11, 5.23447709e-11], [9.18492435e-11, 5.13015949e-11, 5.23350530e-11], [9.18988358e-11, 5.14185847e-11, 5.22869421e-11], ..., [2.72731219e-19, 2.10820705e-19, 1.95528824e-19], [2.77044598e-20, 2.69720236e-20, 2.36522635e-20], [1.77527904e-20, 2.67585028e-20, 2.76238420e-20]], [[9.36112438e-11, 5.22241798e-11, 5.41772079e-11], [9.36148797e-11, 5.22875458e-11, 5.42009007e-11], [9.36260861e-11, 5.24214769e-11, 5.42280006e-11], ..., [2.74349262e-19, 2.30793265e-19, 2.15920196e-19], [2.42248987e-20, 1.72841344e-20, 1.58122392e-20], [1.85720142e-20, 1.72598198e-20, 1.93047783e-20]], [[9.69058306e-11, 5.28302298e-11, 5.56108076e-11], [9.68998146e-11, 5.28920727e-11, 5.56616593e-11], [9.68917169e-11, 5.30219341e-11, 5.57518025e-11], ..., ... ..., [2.42188531e-19, 1.87590604e-19, 1.73665859e-19], [1.86894238e-20, 1.78049916e-20, 1.68037250e-20], [7.37561005e-21, 5.60902813e-21, 5.33276353e-21]], [[8.36587119e-11, 9.20801421e-11, 8.79499112e-11], [8.36090711e-11, 9.21489482e-11, 8.76721681e-11], [8.34909641e-11, 9.23093130e-11, 8.68353514e-11], ..., [2.64613372e-19, 2.26253621e-19, 2.14656678e-19], [1.50624743e-20, 1.47302950e-20, 1.62674519e-20], [6.41543916e-21, 5.28067337e-21, 5.40171720e-21]], [[8.15247037e-11, 1.07296179e-10, 8.96918095e-11], [8.14599360e-11, 1.07303097e-10, 8.92379989e-11], [8.13079673e-11, 1.07308176e-10, 8.79050097e-11], ..., [2.68906620e-19, 2.40782996e-19, 2.24566999e-19], [1.19141053e-20, 1.30351313e-20, 1.44657733e-20], [6.02185911e-21, 5.40972445e-21, 5.67302557e-21]]], dtype=float32) - so4_a2(time, lev, ncol)float322.249e-12 3.721e-12 ... 2.748e-14
- mdims :
- 9
- units :
- kg/kg
- long_name :
- so4_a2 concentration
- basename :
- so4_a2
array([[[2.2489241e-12, 3.7210152e-12, 4.6907001e-12], [2.2477296e-12, 3.7190429e-12, 4.6489353e-12], [2.2451494e-12, 3.7166320e-12, 4.5566866e-12], ..., [4.2287100e-14, 4.1371993e-14, 4.0616168e-14], [3.5077267e-14, 3.2995396e-14, 3.3193693e-14], [3.3943937e-14, 3.2055481e-14, 3.1838915e-14]], [[2.2999660e-12, 3.7788583e-12, 4.8279332e-12], [2.2988605e-12, 3.7759032e-12, 4.7757336e-12], [2.2964172e-12, 3.7721935e-12, 4.6609270e-12], ..., [4.2267703e-14, 4.0747120e-14, 4.0294574e-14], [3.6659986e-14, 3.4782663e-14, 3.5102895e-14], [3.4156695e-14, 3.3951046e-14, 3.3835500e-14]], [[2.3849449e-12, 3.8185470e-12, 4.9417111e-12], [2.3840621e-12, 3.8143941e-12, 4.8796587e-12], [2.3820047e-12, 3.8091019e-12, 4.7483224e-12], ..., ... ..., [3.5025934e-14, 3.4707470e-14, 3.4760199e-14], [3.3869083e-14, 3.3936948e-14, 3.3624694e-14], [3.0480616e-14, 3.0113654e-14, 3.0032976e-14]], [[3.3437732e-12, 4.3878065e-12, 3.9687988e-12], [3.3388513e-12, 4.3886405e-12, 3.9484874e-12], [3.3297265e-12, 4.3927089e-12, 3.8897284e-12], ..., [3.4774541e-14, 3.4977328e-14, 3.5191132e-14], [3.1483479e-14, 3.1627461e-14, 3.2568468e-14], [2.6935695e-14, 2.7354250e-14, 2.8929784e-14]], [[3.3313447e-12, 5.0406901e-12, 3.6872012e-12], [3.3259977e-12, 5.0366738e-12, 3.6549284e-12], [3.3158185e-12, 5.0295141e-12, 3.5623123e-12], ..., [3.4501515e-14, 3.5110403e-14, 3.5525317e-14], [2.8926433e-14, 3.0042548e-14, 3.1647041e-14], [2.4901044e-14, 2.5741718e-14, 2.7477612e-14]]], dtype=float32) - so4_a3(time, lev, ncol)float324.968e-12 8.072e-12 ... 8.885e-32
- mdims :
- 9
- units :
- kg/kg
- long_name :
- so4_a3 concentration
- basename :
- so4_a3
array([[[4.96760098e-12, 8.07165793e-12, 1.02105676e-11], [5.03640098e-12, 8.04598142e-12, 1.04992057e-11], [5.33812144e-12, 7.99075129e-12, 1.06897147e-11], ..., [2.23474535e-28, 3.97702818e-29, 3.41105485e-29], [1.19491599e-31, 1.03909457e-31, 9.25975456e-32], [8.91935902e-32, 8.71327723e-32, 8.85973795e-32]], [[4.90567829e-12, 1.04656075e-11, 1.20657399e-11], [4.96029303e-12, 1.04190484e-11, 1.25074976e-11], [5.22091528e-12, 1.10883013e-11, 1.29871209e-11], ..., [2.06658494e-28, 7.20196507e-29, 5.86557432e-29], [1.10989871e-31, 1.03022511e-31, 8.80473187e-32], [8.49570968e-32, 8.87949155e-32, 9.09262336e-32]], [[1.03958292e-11, 1.01244698e-11, 1.35944138e-11], [1.10314136e-11, 1.24689616e-11, 1.41200220e-11], [1.10173251e-11, 1.42468927e-11, 1.43654004e-11], ..., ... ..., [7.98462629e-29, 5.75878512e-29, 7.05076814e-29], [7.53565759e-32, 7.04493101e-32, 6.55974454e-32], [8.08123743e-32, 8.28156518e-32, 8.71822136e-32]], [[1.71546111e-12, 1.98483873e-12, 2.33115957e-12], [1.73316765e-12, 1.99624258e-12, 2.32121158e-12], [1.72045538e-12, 2.01056121e-12, 2.29228138e-12], ..., [8.24343456e-29, 7.57146135e-29, 8.44169761e-29], [7.32692447e-32, 6.32053908e-32, 6.56208084e-32], [8.46062235e-32, 8.91246710e-32, 8.80086978e-32]], [[2.08404301e-12, 1.72004338e-12, 1.84977142e-12], [2.11411184e-12, 1.72003709e-12, 1.82937910e-12], [2.10541177e-12, 1.71235400e-12, 1.77695337e-12], ..., [7.55454326e-29, 8.16573292e-29, 9.55004923e-29], [6.43600789e-32, 7.54683301e-32, 7.64016139e-32], [8.19624545e-32, 8.91608351e-32, 8.88504282e-32]]], dtype=float32) - so4_c1(time, lev, ncol)float323.976e-20 3.974e-20 ... 3.974e-20
- mdims :
- 9
- units :
- kg/kg
- long_name :
- so4_c1 in cloud water
- basename :
- so4_c1
array([[[3.9757247e-20, 3.9741573e-20, 3.9739185e-20], [3.9745738e-20, 3.9741573e-20, 3.9738833e-20], [3.9741805e-20, 3.9738829e-20, 3.9738778e-20], ..., [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20]], [[3.9757939e-20, 3.9741640e-20, 3.9739110e-20], [3.9745563e-20, 3.9741640e-20, 3.9738816e-20], [3.9741834e-20, 3.9738778e-20, 3.9738778e-20], ..., [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20]], [[3.9738778e-20, 3.9741689e-20, 3.9738962e-20], [3.9764495e-20, 3.9738900e-20, 3.9738778e-20], [3.9747602e-20, 3.9738778e-20, 3.9738778e-20], ..., ... ..., [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20]], [[3.9742497e-20, 3.9739401e-20, 3.9738778e-20], [3.9740468e-20, 3.9739072e-20, 3.9738778e-20], [3.9739282e-20, 3.9738865e-20, 3.9738778e-20], ..., [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20]], [[3.9742707e-20, 3.9740406e-20, 3.9738778e-20], [3.9740535e-20, 3.9739527e-20, 3.9738778e-20], [3.9739295e-20, 3.9738997e-20, 3.9738778e-20], ..., [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20]]], dtype=float32) - so4_c2(time, lev, ncol)float323.972e-20 3.974e-20 ... 3.974e-20
- mdims :
- 9
- units :
- kg/kg
- long_name :
- so4_c2 in cloud water
- basename :
- so4_c2
array([[[3.9720305e-20, 3.9735983e-20, 3.9738367e-20], [3.9731814e-20, 3.9735979e-20, 3.9738720e-20], [3.9735750e-20, 3.9738723e-20, 3.9738778e-20], ..., [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20]], [[3.9719617e-20, 3.9735912e-20, 3.9738445e-20], [3.9731989e-20, 3.9735912e-20, 3.9738736e-20], [3.9735718e-20, 3.9738778e-20, 3.9738778e-20], ..., [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20]], [[3.9738778e-20, 3.9735863e-20, 3.9738590e-20], [3.9713058e-20, 3.9738652e-20, 3.9738778e-20], [3.9729953e-20, 3.9738778e-20, 3.9738778e-20], ..., ... ..., [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20]], [[3.9735055e-20, 3.9738151e-20, 3.9738778e-20], [3.9737085e-20, 3.9738484e-20, 3.9738778e-20], [3.9738270e-20, 3.9738687e-20, 3.9738778e-20], ..., [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20]], [[3.9734845e-20, 3.9737146e-20, 3.9738778e-20], [3.9737017e-20, 3.9738025e-20, 3.9738778e-20], [3.9738257e-20, 3.9738558e-20, 3.9738778e-20], ..., [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20]]], dtype=float32) - so4_c3(time, lev, ncol)float323.974e-20 3.974e-20 ... 3.974e-20
- mdims :
- 9
- units :
- kg/kg
- long_name :
- so4_c3 in cloud water
- basename :
- so4_c3
array([[[3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], ..., [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20]], [[3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], ..., [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20]], [[3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], ..., ... ..., [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20]], [[3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], ..., [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20]], [[3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], ..., [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20], [3.9738778e-20, 3.9738778e-20, 3.9738778e-20]]], dtype=float32) - soa_a1(time, lev, ncol)float328.4e-11 7.284e-11 ... 3.894e-22
- mdims :
- 9
- units :
- kg/kg
- long_name :
- soa_a1 concentration
- basename :
- soa_a1
array([[[8.39987940e-11, 7.28385477e-11, 8.92462770e-11], [8.40596759e-11, 7.28995336e-11, 8.92070237e-11], [8.41819045e-11, 7.30185981e-11, 8.90465410e-11], ..., [1.86000043e-20, 1.55599814e-20, 1.44535336e-20], [1.57508457e-21, 1.38010582e-21, 1.22293978e-21], [1.01183122e-21, 1.25151387e-21, 1.28098924e-21]], [[8.52423063e-11, 6.77903844e-11, 8.58280252e-11], [8.52618531e-11, 6.78543610e-11, 8.58208296e-11], [8.53087045e-11, 6.79748063e-11, 8.57209581e-11], ..., [1.86927535e-20, 1.74370707e-20, 1.65223511e-20], [1.31364985e-21, 8.84414200e-22, 8.18326236e-22], [1.08124148e-21, 8.50741628e-22, 8.85763922e-22]], [[8.93157562e-11, 6.31248803e-11, 8.22992438e-11], [8.93066246e-11, 6.31849503e-11, 8.23212887e-11], [8.93003380e-11, 6.32915248e-11, 8.22720780e-11], ..., ... ..., [2.15252671e-20, 1.64385328e-20, 1.53032307e-20], [1.20755556e-21, 1.22691231e-21, 1.23088433e-21], [4.67875145e-22, 3.69055839e-22, 3.27735961e-22]], [[2.93856918e-11, 1.11497797e-10, 1.21577567e-10], [2.94312422e-11, 1.11508164e-10, 1.21353705e-10], [2.95043642e-11, 1.11502370e-10, 1.20652405e-10], ..., [2.35814103e-20, 1.98627698e-20, 1.88478967e-20], [1.13934204e-21, 1.14563202e-21, 1.22359258e-21], [4.72183461e-22, 3.74419134e-22, 3.60824372e-22]], [[2.50606445e-11, 9.52140519e-11, 1.01384977e-10], [2.51104224e-11, 9.51441287e-11, 1.00961968e-10], [2.51959478e-11, 9.49383350e-11, 9.96863148e-11], ..., [2.40841309e-20, 2.10405683e-20, 1.95810114e-20], [9.71692357e-22, 1.01024178e-21, 1.08019509e-21], [4.82148706e-22, 4.00079504e-22, 3.89427383e-22]]], dtype=float32) - soa_a2(time, lev, ncol)float324.754e-13 6.361e-13 ... 1.923e-16
- mdims :
- 9
- units :
- kg/kg
- long_name :
- soa_a2 concentration
- basename :
- soa_a2
array([[[4.7541403e-13, 6.3606764e-13, 9.3555144e-13], [4.7819810e-13, 6.3784931e-13, 9.3658100e-13], [4.8368449e-13, 6.4052051e-13, 9.3664193e-13], ..., [4.1459327e-16, 3.8865411e-16, 3.6869290e-16], [2.7734572e-16, 2.4715728e-16, 2.4648709e-16], [2.6725300e-16, 2.3024799e-16, 2.2471390e-16]], [[4.8255139e-13, 5.4816546e-13, 8.5231317e-13], [4.8462184e-13, 5.5031630e-13, 8.5345880e-13], [4.8878940e-13, 5.5324126e-13, 8.5284915e-13], ..., [3.9816960e-16, 3.8309686e-16, 3.7138742e-16], [2.9408383e-16, 2.6349946e-16, 2.6505921e-16], [2.7131405e-16, 2.5050613e-16, 2.4292299e-16]], [[5.2034820e-13, 4.8415102e-13, 7.8871420e-13], [5.2180021e-13, 4.8640969e-13, 7.8981830e-13], [5.2479993e-13, 4.8890915e-13, 7.8661535e-13], ..., ... ..., [2.7323864e-16, 2.6547402e-16, 2.6527158e-16], [2.4476732e-16, 2.4322458e-16, 2.3998463e-16], [2.0160779e-16, 1.9638925e-16, 1.9729151e-16]], [[2.0208974e-13, 1.2944174e-12, 1.7207035e-12], [2.0881223e-13, 1.2945054e-12, 1.7210130e-12], [2.2127115e-13, 1.2929396e-12, 1.7201555e-12], ..., [2.7845163e-16, 2.7685592e-16, 2.7624754e-16], [2.2796513e-16, 2.2733086e-16, 2.3123412e-16], [1.9063636e-16, 1.9029523e-16, 1.9468123e-16]], [[2.3227015e-13, 8.6460711e-13, 1.3332078e-12], [2.3904007e-13, 8.6507640e-13, 1.3293448e-12], [2.5177714e-13, 8.6392747e-13, 1.3160651e-12], ..., [2.7632163e-16, 2.7887586e-16, 2.8008584e-16], [2.1385750e-16, 2.1907673e-16, 2.2444970e-16], [1.8369887e-16, 1.8581409e-16, 1.9227365e-16]]], dtype=float32) - soa_a3(time, lev, ncol)float325.266e-12 1.384e-11 ... 1.005e-36
- mdims :
- 9
- units :
- kg/kg
- long_name :
- soa_a3 concentration
- basename :
- soa_a3
array([[[5.26648690e-12, 1.38420777e-11, 1.95408550e-11], [5.32790955e-12, 1.38078125e-11, 2.01121324e-11], [5.59568798e-12, 1.37326929e-11, 2.04205056e-11], ..., [1.00000004e-36, 1.00012559e-36, 1.00008766e-36], [1.00050558e-36, 1.00000004e-36, 1.00000004e-36], [1.00000004e-36, 1.00814192e-36, 1.01708601e-36]], [[5.17024141e-12, 1.62146459e-11, 2.13967993e-11], [5.21737211e-12, 1.61559655e-11, 2.21567105e-11], [5.44431817e-12, 1.69610333e-11, 2.28427641e-11], ..., [1.00003950e-36, 1.00013797e-36, 1.00000004e-36], [1.00019653e-36, 1.00000004e-36, 1.00000004e-36], [1.00000004e-36, 1.00638619e-36, 1.02306483e-36]], [[1.00755550e-11, 1.50144931e-11, 2.28015263e-11], [1.06348845e-11, 1.76577503e-11, 2.37203625e-11], [1.06198219e-11, 1.96552011e-11, 2.40451947e-11], ..., ... ..., [1.00000004e-36, 1.00000004e-36, 1.00000004e-36], [1.00029940e-36, 1.00000004e-36, 1.00000004e-36], [1.00000004e-36, 1.00746203e-36, 1.00878010e-36]], [[1.84019098e-12, 4.36502093e-12, 5.32845426e-12], [1.86121908e-12, 4.37660541e-12, 5.31239679e-12], [1.86918580e-12, 4.39031754e-12, 5.26438051e-12], ..., [1.00003250e-36, 1.00000004e-36, 1.00000004e-36], [1.00014326e-36, 1.00006524e-36, 1.00000004e-36], [1.00229852e-36, 1.00456930e-36, 1.00836855e-36]], [[2.11200762e-12, 3.46470530e-12, 4.11106088e-12], [2.14162585e-12, 3.46445875e-12, 4.07655984e-12], [2.15204850e-12, 3.45478984e-12, 3.98181878e-12], ..., [1.00000004e-36, 1.00007663e-36, 1.00000004e-36], [1.00000004e-36, 1.00063652e-36, 1.00000004e-36], [1.00562415e-36, 1.00114215e-36, 1.00483189e-36]]], dtype=float32) - soa_c1(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- soa_c1 in cloud water
- basename :
- soa_c1
array([[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 1.4165412e-26, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [1.8238289e-27, 6.4538818e-27, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - soa_c2(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- soa_c2 in cloud water
- basename :
- soa_c2
array([[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 1.4640563e-28, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [1.0368731e-29, 5.2153942e-29, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - soa_c3(time, lev, ncol)float320.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- mdims :
- 9
- units :
- kg/kg
- long_name :
- soa_c3 in cloud water
- basename :
- soa_c3
array([[[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 5.4803179e-28, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [1.4331882e-28, 2.3316926e-28, 0.0000000e+00], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - wat_a1(time, lev, ncol)float323.214e-10 1.53e-10 ... 0.0 0.0
- mdims :
- 9
- units :
- m
- long_name :
- aerosol water, interstitial, mode 01
- basename :
- wat_a1
array([[[3.2135011e-10, 1.5303454e-10, 2.9589073e-10], [3.5732087e-10, 1.7077792e-10, 3.0969785e-10], [4.4127707e-10, 2.0174584e-10, 3.3870806e-10], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[3.0491307e-10, 1.6085513e-10, 3.0302041e-10], [3.3728759e-10, 1.8037412e-10, 3.1816569e-10], [4.1163994e-10, 2.1291417e-10, 3.4603473e-10], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[2.9162270e-10, 1.6901690e-10, 3.1118697e-10], [3.2130118e-10, 1.9065520e-10, 3.2824324e-10], [3.8799089e-10, 2.2271385e-10, 3.4598710e-10], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[7.7395779e-10, 7.8822315e-10, 5.9329708e-10], [8.8015772e-10, 9.3212471e-10, 6.8736827e-10], [1.1050580e-09, 1.2474981e-09, 9.0682289e-10], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[7.4695328e-10, 8.7925173e-10, 6.1191863e-10], [8.5111573e-10, 1.0477544e-09, 7.2556222e-10], [1.0747017e-09, 1.4341347e-09, 1.0189177e-09], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - wat_a2(time, lev, ncol)float323.821e-12 3.4e-12 ... 0.0 0.0
- mdims :
- 9
- units :
- m
- long_name :
- aerosol water, interstitial, mode 02
- basename :
- wat_a2
array([[[3.82058838e-12, 3.39982708e-12, 7.28252614e-12], [4.22146736e-12, 3.78251250e-12, 7.55945393e-12], [5.13985165e-12, 4.44582912e-12, 8.12025360e-12], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[3.65510747e-12, 3.64645770e-12, 7.63812798e-12], [4.02018176e-12, 4.07456577e-12, 7.93614306e-12], [4.84438401e-12, 4.78237762e-12, 8.44144980e-12], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[3.52938252e-12, 3.92898821e-12, 8.04495793e-12], [3.86859642e-12, 4.41373023e-12, 8.37814193e-12], [4.61898792e-12, 5.12479425e-12, 8.61862491e-12], ..., ... ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[9.97772438e-12, 1.19765317e-11, 1.05667029e-11], [1.12192061e-11, 1.39774121e-11, 1.20726354e-11], [1.37698802e-11, 1.82159010e-11, 1.54331756e-11], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[9.77830317e-12, 1.34120874e-11, 1.02203298e-11], [1.10152373e-11, 1.57575706e-11, 1.18876567e-11], [1.35902513e-11, 2.09342578e-11, 1.59082001e-11], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]]], dtype=float32) - wat_a3(time, lev, ncol)float322.606e-09 2.395e-09 ... 0.0 0.0
- mdims :
- 9
- units :
- m
- long_name :
- aerosol water, interstitial, mode 03
- basename :
- wat_a3
array([[[2.6057103e-09, 2.3951992e-09, 4.6254702e-09], [2.8976659e-09, 2.6716862e-09, 4.8354991e-09], [3.5829792e-09, 3.1510736e-09, 5.2732632e-09], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[2.2657052e-09, 2.5631288e-09, 4.7639306e-09], [2.5066720e-09, 2.8724219e-09, 5.0002131e-09], [3.0632161e-09, 3.3834264e-09, 5.4309184e-09], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[2.0048059e-09, 2.6244202e-09, 4.8516107e-09], [2.2079187e-09, 2.9593146e-09, 5.1094040e-09], [2.6668405e-09, 3.4498784e-09, 5.3558189e-09], ..., ... ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[9.6626209e-09, 3.2703031e-09, 3.7277013e-09], [1.1005202e-08, 3.8877208e-09, 4.3424060e-09], [1.3851345e-08, 5.2589666e-09, 5.7985816e-09], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]], [[9.6572030e-09, 3.6618437e-09, 3.5761822e-09], [1.1020516e-08, 4.3862340e-09, 4.2635810e-09], [1.3951010e-08, 6.0695613e-09, 6.0637992e-09], ..., [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00], [0.0000000e+00, 0.0000000e+00, 0.0000000e+00]]], dtype=float32) - wat_a4(time, lev, ncol)float321.733e-17 9.715e-18 ... 0.0 0.0
- mdims :
- 9
- units :
- m
- long_name :
- aerosol water, interstitial, mode 04
- basename :
- wat_a4
array([[[1.73314604e-17, 9.71464321e-18, 1.29712318e-17], [1.91857711e-17, 1.08254161e-17, 1.35958747e-17], [2.34209838e-17, 1.27612048e-17, 1.49277629e-17], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[1.68535767e-17, 1.11682411e-17, 1.47178724e-17], [1.85725076e-17, 1.25015404e-17, 1.54761588e-17], [2.24437724e-17, 1.47207428e-17, 1.69131254e-17], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[1.60673680e-17, 1.27436272e-17, 1.66056276e-17], [1.76455938e-17, 1.43449447e-17, 1.75354994e-17], [2.11305074e-17, 1.67168702e-17, 1.86060035e-17], ..., ... ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[5.52600390e-17, 5.13061885e-17, 4.80779474e-17], [6.24388617e-17, 5.98489525e-17, 5.52216049e-17], [7.73621859e-17, 7.77397575e-17, 7.15856727e-17], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[6.67395920e-17, 5.36767029e-17, 5.52520650e-17], [7.55542867e-17, 6.30174513e-17, 6.48441044e-17], [9.41079214e-17, 8.33651680e-17, 8.88307407e-17], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]]], dtype=float32) - p_3d(time, lev, ncol)float641.033e+03 1.029e+03 ... 0.1238
- units :
- hPa
array([[[1.03291446e+03, 1.02869331e+03, 1.02154642e+03], [1.02801160e+03, 1.02381050e+03, 1.01669753e+03], [1.02018710e+03, 1.01601946e+03, 1.00896315e+03], ..., [2.69948862e-01, 2.69948862e-01, 2.69948862e-01], [1.82829236e-01, 1.82829236e-01, 1.82829236e-01], [1.23825413e-01, 1.23825413e-01, 1.23825413e-01]], [[1.03322422e+03, 1.02919693e+03, 1.02209699e+03], [1.02831990e+03, 1.02431172e+03, 1.01724549e+03], [1.02049294e+03, 1.01651669e+03, 1.00950675e+03], ..., [2.69948862e-01, 2.69948862e-01, 2.69948862e-01], [1.82829236e-01, 1.82829236e-01, 1.82829236e-01], [1.23825413e-01, 1.23825413e-01, 1.23825413e-01]], [[1.03353243e+03, 1.02975710e+03, 1.02270132e+03], [1.02862665e+03, 1.02486924e+03, 1.01784694e+03], [1.02079724e+03, 1.01706977e+03, 1.01010342e+03], ..., ... ..., [2.69948862e-01, 2.69948862e-01, 2.69948862e-01], [1.82829236e-01, 1.82829236e-01, 1.82829236e-01], [1.23825413e-01, 1.23825413e-01, 1.23825413e-01]], [[1.01771079e+03, 1.02224919e+03, 1.01778684e+03], [1.01288010e+03, 1.01739696e+03, 1.01295580e+03], [1.00517614e+03, 1.00965702e+03, 1.00525123e+03], ..., [2.69948862e-01, 2.69948862e-01, 2.69948862e-01], [1.82829236e-01, 1.82829236e-01, 1.82829236e-01], [1.23825413e-01, 1.23825413e-01, 1.23825413e-01]], [[1.01721895e+03, 1.02088117e+03, 1.01658342e+03], [1.01239060e+03, 1.01603543e+03, 1.01175809e+03], [1.00469054e+03, 1.00830634e+03, 1.00406306e+03], ..., [2.69948862e-01, 2.69948862e-01, 2.69948862e-01], [1.82829236e-01, 1.82829236e-01, 1.82829236e-01], [1.23825413e-01, 1.23825413e-01, 1.23825413e-01]]]) - zeros_cf(time, lev, ncol)float640.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- long_name :
- An array of zeros as only strat output is used for this model
array([[[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., ... ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]]]) - rho_a(time, lev, ncol)float641.348 1.336 ... 0.0001633 0.0001627
- units :
- kg / m ** 3
array([[[1.34772158e+00, 1.33567682e+00, 1.33317273e+00], [1.34320972e+00, 1.33116818e+00, 1.32823899e+00], [1.33598602e+00, 1.32400278e+00, 1.32066819e+00], ..., [3.62402305e-04, 3.60603177e-04, 3.59287195e-04], [2.39792287e-04, 2.36321634e-04, 2.35348150e-04], [1.62939565e-04, 1.60291213e-04, 1.59456697e-04]], [[1.34767338e+00, 1.33570042e+00, 1.33344234e+00], [1.34314474e+00, 1.33119935e+00, 1.32846552e+00], [1.33589492e+00, 1.32405682e+00, 1.32086051e+00], ..., [3.61644353e-04, 3.60988346e-04, 3.59847140e-04], [2.38122971e-04, 2.34533173e-04, 2.33675512e-04], [1.62581920e-04, 1.59380998e-04, 1.58550150e-04]], [[1.34762880e+00, 1.33583878e+00, 1.33372085e+00], [1.34308079e+00, 1.33135981e+00, 1.32874153e+00], [1.33580583e+00, 1.32421193e+00, 1.32113585e+00], ..., ... ..., [3.71647061e-04, 3.68894295e-04, 3.67580398e-04], [2.49620214e-04, 2.48576785e-04, 2.47830592e-04], [1.65775898e-04, 1.63928717e-04, 1.63401397e-04]], [[1.30265795e+00, 1.32040925e+00, 1.32878978e+00], [1.29823239e+00, 1.31598821e+00, 1.32411806e+00], [1.29114861e+00, 1.30889945e+00, 1.31650406e+00], ..., [3.71380151e-04, 3.68906662e-04, 3.67659326e-04], [2.48438586e-04, 2.46832940e-04, 2.46194172e-04], [1.65566379e-04, 1.63688414e-04, 1.63396108e-04]], [[1.30264403e+00, 1.31614041e+00, 1.32324088e+00], [1.29822494e+00, 1.31172804e+00, 1.31861667e+00], [1.29115521e+00, 1.30463766e+00, 1.31110929e+00], ..., [3.70056960e-04, 3.67430108e-04, 3.66223381e-04], [2.46701977e-04, 2.44524482e-04, 2.43824965e-04], [1.65559720e-04, 1.63332864e-04, 1.62722915e-04]]]) - strat_frac_subcolumns_cl(subcolumn, time, lev, ncol)boolFalse False False ... False False
- long_name :
- Liquid cloud particles present? [stratiform]
- units :
- 0 = no, 1 = yes
- Processing method :
- Radiation logic
array([[[[False, False, False], [False, False, False], [False, False, False], ..., [False, False, False], [False, False, False], [False, False, False]], [[False, False, False], [False, False, False], [False, False, False], ..., [False, False, False], [False, False, False], [False, False, False]], [[False, False, False], [False, False, False], [False, False, False], ..., ... ..., [False, False, False], [False, False, False], [False, False, False]], [[False, False, False], [False, False, False], [False, False, False], ..., [False, False, False], [False, False, False], [False, False, False]], [[False, False, False], [False, False, False], [False, False, False], ..., [False, False, False], [False, False, False], [False, False, False]]]]) - strat_frac_subcolumns_ci(subcolumn, time, lev, ncol)boolFalse False False ... False False
- long_name :
- Liquid cloud particles present? [stratiform]
- units :
- 0 = no, 1 = yes
- Processing method :
- Radiation logic
array([[[[False, False, False], [False, False, False], [False, False, False], ..., [False, False, False], [False, False, False], [False, False, False]], [[False, False, False], [False, False, False], [False, False, False], ..., [False, False, False], [False, False, False], [False, False, False]], [[False, False, False], [False, False, False], [False, False, False], ..., ... ..., [False, False, False], [False, False, False], [False, False, False]], [[False, False, False], [False, False, False], [False, False, False], ..., [False, False, False], [False, False, False], [False, False, False]], [[False, False, False], [False, False, False], [False, False, False], ..., [False, False, False], [False, False, False], [False, False, False]]]]) - strat_frac_subcolumns_pl(subcolumn, time, lev, ncol)boolFalse False False ... False False
- long_name :
- Liquid precipitation present? [stratiform]
- units :
- 0 = no, 1 = yes
- Processing method :
- Radiation logic
array([[[[False, False, False], [False, False, False], [False, False, False], ..., [False, False, False], [False, False, False], [False, False, False]], [[False, False, False], [False, False, False], [False, False, False], ..., [False, False, False], [False, False, False], [False, False, False]], [[False, False, False], [False, False, False], [False, False, False], ..., ... ..., [False, False, False], [False, False, False], [False, False, False]], [[False, False, False], [False, False, False], [False, False, False], ..., [False, False, False], [False, False, False], [False, False, False]], [[False, False, False], [False, False, False], [False, False, False], ..., [False, False, False], [False, False, False], [False, False, False]]]]) - strat_frac_subcolumns_pi(subcolumn, time, lev, ncol)boolFalse False False ... False False
- long_name :
- Ice precipitation present? [stratiform]
- units :
- 0 = no, 1 = yes
- Processing method :
- Radiation logic
array([[[[False, False, False], [False, False, False], [False, False, False], ..., [False, False, False], [False, False, False], [False, False, False]], [[False, False, False], [False, False, False], [False, False, False], ..., [False, False, False], [False, False, False], [False, False, False]], [[False, False, False], [False, False, False], [False, False, False], ..., ... ..., [False, False, False], [False, False, False], [False, False, False]], [[False, False, False], [False, False, False], [False, False, False], ..., [False, False, False], [False, False, False], [False, False, False]], [[False, False, False], [False, False, False], [False, False, False], ..., [False, False, False], [False, False, False], [False, False, False]]]]) - strat_q_subcolumns_cl(subcolumn, time, lev, ncol)float640.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- long_name :
- q in subcolumns
- units :
- $kg\ kg^{-1}$
- Processing method :
- Radiation logic
array([[[[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., ... ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]]]]) - strat_n_subcolumns_cl(subcolumn, time, lev, ncol)float640.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- long_name :
- N in subcolumns
- units :
- $cm^{-3}$
- Processing method :
- Radiation logic
array([[[[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., ... ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]]]]) - strat_q_subcolumns_ci(subcolumn, time, lev, ncol)float640.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- long_name :
- q in subcolumns
- units :
- $kg\ kg^{-1}$
- Processing method :
- Radiation logic
array([[[[0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], ..., ... ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]]]]) - strat_n_subcolumns_ci(subcolumn, time, lev, ncol)float640.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- long_name :
- N in subcolumns
- units :
- $cm^{-3}$
- Processing method :
- Radiation logic
array([[[[0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], ..., ... ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]], [[0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00]]]]) - strat_q_subcolumns_pl(subcolumn, time, lev, ncol)float640.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- long_name :
- q in subcolumns
- units :
- $kg\ kg^{-1}$
- Processing method :
- Radiation logic
array([[[[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., ... ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]]]]) - strat_n_subcolumns_pl(subcolumn, time, lev, ncol)float640.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- long_name :
- N in subcolumns
- units :
- $cm^{-3}$
- Processing method :
- Radiation logic
array([[[[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., ... ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]]]]) - strat_q_subcolumns_pi(subcolumn, time, lev, ncol)float640.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- long_name :
- q in subcolumns
- units :
- $kg\ kg^{-1}$
- Processing method :
- Radiation logic
array([[[[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., ... ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]]]]) - strat_n_subcolumns_pi(subcolumn, time, lev, ncol)float640.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- long_name :
- N in subcolumns
- units :
- $cm^{-3}$
- Processing method :
- Radiation logic
array([[[[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., ... ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [0., 0., 0.], [0., 0., 0.], [0., 0., 0.]]]]) - tau(time, lev, ncol)float641.0 1.0 1.0 ... 0.798 0.7973 0.7981
- long_name :
- Two-way transmittance
- units :
- 1
array([[[1. , 1. , 1. ], [0.99891197, 0.99891642, 0.99892389], [0.99717842, 0.9971903 , 0.99721025], ..., [0.79527422, 0.79601749, 0.79728284], [0.79525883, 0.79600207, 0.79726739], [0.79524844, 0.79599167, 0.79725697]], [[1. , 1. , 1. ], [0.99891165, 0.99891586, 0.99892328], [0.99717755, 0.9971888 , 0.99720864], ..., [0.79521928, 0.79592786, 0.79718494], [0.79520388, 0.79591243, 0.79716948], [0.79519349, 0.79590202, 0.79715906]], [[1. , 1. , 1. ], [0.99891132, 0.99891524, 0.99892262], [0.99717669, 0.99718715, 0.99720688], ..., ... ..., [0.79775631, 0.79684811, 0.79767747], [0.7977409 , 0.79683272, 0.79766206], [0.79773045, 0.79682228, 0.79765161]], [[1. , 1. , 1. ], [0.99892729, 0.9989228 , 0.99892778], [0.99721931, 0.99720733, 0.99722057], ..., [0.79790607, 0.797114 , 0.7979074 ], [0.79789065, 0.7970986 , 0.79789198], [0.7978802 , 0.79708815, 0.79788153]], [[1. , 1. , 1. ], [0.99892782, 0.99892417, 0.99892895], [0.99722073, 0.99721096, 0.99722369], ..., [0.79799496, 0.79735303, 0.79811713], [0.79797953, 0.79733761, 0.79810169], [0.79796909, 0.79732718, 0.79809124]]]) - u(time, lev, ncol)float640.0 0.0 0.0 ... 0.1132 0.1128
- long_name :
- Atmospheric optical depth
- units :
- 1
array([[[0. , 0. , 0. ], [0.00054431, 0.00054208, 0.00053835], [0.00141278, 0.00140683, 0.00139682], ..., [0.11453415, 0.11406706, 0.11327289], [0.11454382, 0.11407675, 0.11328258], [0.11455035, 0.11408328, 0.11328912]], [[0. , 0. , 0. ], [0.00054447, 0.00054236, 0.00053865], [0.00141322, 0.00140758, 0.00139763], ..., [0.11456869, 0.11412336, 0.11333429], [0.11457837, 0.11413306, 0.11334399], [0.1145849 , 0.11413959, 0.11335052]], [[0. , 0. , 0. ], [0.00054464, 0.00054267, 0.00053898], [0.00141365, 0.00140841, 0.00139851], ..., ... ..., [0.11297605, 0.1135456 , 0.11302547], [0.11298571, 0.11355525, 0.11303512], [0.11299226, 0.11356181, 0.11304168]], [[0. , 0. , 0. ], [0.00053664, 0.00053889, 0.0005364 ], [0.00139228, 0.00139829, 0.00139165], ..., [0.1128822 , 0.11337879, 0.11288136], [0.11289186, 0.11338845, 0.11289103], [0.11289841, 0.113395 , 0.11289758]], [[0. , 0. , 0. ], [0.00053638, 0.00053821, 0.00053581], [0.00139157, 0.00139647, 0.00139009], ..., [0.1128265 , 0.11322887, 0.11274996], [0.11283617, 0.11323854, 0.11275963], [0.11284271, 0.11324509, 0.11276617]]]) - beta(time, lev, ncol)float641.465e-05 1.452e-05 ... 1.768e-09
- long_name :
- Volume extinction cross section
- units :
- m-1
array([[[1.46470472e-05, 1.45161450e-05, 1.44888820e-05], [1.45979842e-05, 1.44671206e-05, 1.44352374e-05], [1.45194368e-05, 1.43892044e-05, 1.43529183e-05], ..., [3.93732255e-09, 3.91778772e-09, 3.90349848e-09], [2.60525936e-09, 2.56757267e-09, 2.55700177e-09], [1.77027837e-09, 1.74152072e-09, 1.73245865e-09]], [[1.46465249e-05, 1.45164036e-05, 1.44918157e-05], [1.45972839e-05, 1.44674610e-05, 1.44377010e-05], [1.45184462e-05, 1.43897949e-05, 1.43550135e-05], ..., [3.92909342e-09, 3.92196921e-09, 3.90957854e-09], [2.58713313e-09, 2.54815225e-09, 2.53883875e-09], [1.76639515e-09, 1.73163693e-09, 1.72261456e-09]], [[1.46460439e-05, 1.45179134e-05, 1.44948452e-05], [1.45965918e-05, 1.44692106e-05, 1.44407053e-05], [1.45174816e-05, 1.43914849e-05, 1.43580078e-05], ..., ... ..., [4.03770300e-09, 4.00781455e-09, 3.99354753e-09], [2.71197465e-09, 2.70064431e-09, 2.69254234e-09], [1.80107676e-09, 1.78101874e-09, 1.77529307e-09]], [[1.41573011e-05, 1.43502072e-05, 1.44412311e-05], [1.41091805e-05, 1.43021334e-05, 1.43904321e-05], [1.40321511e-05, 1.42250542e-05, 1.43076443e-05], ..., [4.03480601e-09, 4.00794877e-09, 3.99440455e-09], [2.69914405e-09, 2.68170959e-09, 2.67477412e-09], [1.79880145e-09, 1.77840957e-09, 1.77523551e-09]], [[1.41571458e-05, 1.43038159e-05, 1.43809280e-05], [1.41090923e-05, 1.42558365e-05, 1.43306520e-05], [1.40322201e-05, 1.41787371e-05, 1.42490212e-05], ..., [4.02043792e-09, 3.99191493e-09, 3.97881322e-09], [2.68028770e-09, 2.65664354e-09, 2.64904841e-09], [1.79872898e-09, 1.77454895e-09, 1.76792577e-09]]]) - sigma_180_vol(time, lev, ncol)float641.718e-06 1.703e-06 ... 2.074e-10
- long_name :
- Volume backscatter cross section
- units :
- $m^{-1}$
array([[[1.71796754e-06, 1.70261388e-06, 1.69941618e-06], [1.71221289e-06, 1.69686376e-06, 1.69312414e-06], [1.70299998e-06, 1.68772489e-06, 1.68346886e-06], ..., [4.61812695e-10, 4.59521434e-10, 4.57845434e-10], [3.05573605e-10, 3.01153293e-10, 2.99913422e-10], [2.07637808e-10, 2.04264794e-10, 2.03201893e-10]], [[1.71790628e-06, 1.70264421e-06, 1.69976027e-06], [1.71213075e-06, 1.69690369e-06, 1.69341311e-06], [1.70288380e-06, 1.68779415e-06, 1.68371460e-06], ..., [4.60847492e-10, 4.60011885e-10, 4.58558571e-10], [3.03447560e-10, 2.98875452e-10, 2.97783062e-10], [2.07182342e-10, 2.03105513e-10, 2.02047270e-10]], [[1.71784986e-06, 1.70282131e-06, 1.70011561e-06], [1.71204958e-06, 1.69710890e-06, 1.69376548e-06], [1.70277065e-06, 1.68799237e-06, 1.68406580e-06], ..., ... ..., [4.73586423e-10, 4.70080775e-10, 4.68407382e-10], [3.18090353e-10, 3.16761406e-10, 3.15811118e-10], [2.11250183e-10, 2.08897558e-10, 2.08225988e-10]], [[1.66052471e-06, 1.68315087e-06, 1.69382716e-06], [1.65488059e-06, 1.67751224e-06, 1.68786889e-06], [1.64584573e-06, 1.66847153e-06, 1.67815862e-06], ..., [4.73246632e-10, 4.70096518e-10, 4.68507902e-10], [3.16585439e-10, 3.14540533e-10, 3.13727064e-10], [2.10983311e-10, 2.08591525e-10, 2.08219236e-10]], [[1.66050650e-06, 1.67770958e-06, 1.68675414e-06], [1.65487025e-06, 1.67208202e-06, 1.68085721e-06], [1.64585382e-06, 1.66303896e-06, 1.67128265e-06], ..., [4.71561384e-10, 4.68215892e-10, 4.66679179e-10], [3.14373758e-10, 3.11600510e-10, 3.10709668e-10], [2.10974811e-10, 2.08138709e-10, 2.07361869e-10]]]) - sigma_180(time, lev, ncol)float646.131e-32 6.131e-32 ... 6.129e-32
- long_name :
- backscatter cross section per molecule
- units :
- $m^2$
array([[[6.13095614e-32, 6.13095671e-32, 6.13093590e-32], [6.13094431e-32, 6.13094640e-32, 6.13092529e-32], [6.13092741e-32, 6.13092806e-32, 6.13090842e-32], ..., [6.12898191e-32, 6.12900057e-32, 6.12901358e-32], [6.12906287e-32, 6.12911227e-32, 6.12912600e-32], [6.12905244e-32, 6.12910757e-32, 6.12912485e-32]], [[6.13095670e-32, 6.13095711e-32, 6.13093772e-32], [6.13094715e-32, 6.13094688e-32, 6.13092599e-32], [6.13092724e-32, 6.13092893e-32, 6.13091067e-32], ..., [6.12899111e-32, 6.12899641e-32, 6.12900843e-32], [6.12908722e-32, 6.12913817e-32, 6.12914949e-32], [6.12906103e-32, 6.12912700e-32, 6.12914362e-32]], [[6.13095801e-32, 6.13096028e-32, 6.13093843e-32], [6.13094838e-32, 6.13094942e-32, 6.13092786e-32], [6.13092872e-32, 6.13093061e-32, 6.13091127e-32], ..., ... ..., [6.12889214e-32, 6.12892075e-32, 6.12893309e-32], [6.12892331e-32, 6.12893686e-32, 6.12894839e-32], [6.12899381e-32, 6.12903049e-32, 6.12904242e-32]], [[6.13095907e-32, 6.13095254e-32, 6.13092820e-32], [6.13094868e-32, 6.13094149e-32, 6.13091759e-32], [6.13092950e-32, 6.13092468e-32, 6.13090023e-32], ..., [6.12889692e-32, 6.12892057e-32, 6.12893123e-32], [6.12893960e-32, 6.12896283e-32, 6.12897232e-32], [6.12899723e-32, 6.12903615e-32, 6.12904207e-32]], [[6.13095692e-32, 6.13095345e-32, 6.13092949e-32], [6.13094567e-32, 6.13094250e-32, 6.13092039e-32], [6.13092910e-32, 6.13092450e-32, 6.13090309e-32], ..., [6.12890854e-32, 6.12893287e-32, 6.12894580e-32], [6.12896438e-32, 6.12899503e-32, 6.12900623e-32], [6.12899621e-32, 6.12904449e-32, 6.12905684e-32]]]) - sigma(time, lev, ncol)float645.227e-31 5.227e-31 ... 5.226e-31
- long_name :
- Rayleigh scattering cross section per molecule
- units :
- $m^2$
array([[[5.22713042e-31, 5.22713091e-31, 5.22711317e-31], [5.22712034e-31, 5.22712212e-31, 5.22710412e-31], [5.22710592e-31, 5.22710648e-31, 5.22708974e-31], ..., [5.22544724e-31, 5.22546314e-31, 5.22547424e-31], [5.22551625e-31, 5.22555837e-31, 5.22557008e-31], [5.22550737e-31, 5.22555437e-31, 5.22556910e-31]], [[5.22713090e-31, 5.22713125e-31, 5.22711472e-31], [5.22712276e-31, 5.22712253e-31, 5.22710471e-31], [5.22710578e-31, 5.22710722e-31, 5.22709166e-31], ..., [5.22545508e-31, 5.22545959e-31, 5.22546984e-31], [5.22553702e-31, 5.22558045e-31, 5.22559011e-31], [5.22551469e-31, 5.22557093e-31, 5.22558510e-31]], [[5.22713202e-31, 5.22713395e-31, 5.22711532e-31], [5.22712380e-31, 5.22712469e-31, 5.22710631e-31], [5.22710705e-31, 5.22710866e-31, 5.22709216e-31], ..., ... ..., [5.22537069e-31, 5.22539509e-31, 5.22540561e-31], [5.22539728e-31, 5.22540882e-31, 5.22541865e-31], [5.22545738e-31, 5.22548865e-31, 5.22549883e-31]], [[5.22713292e-31, 5.22712735e-31, 5.22710660e-31], [5.22712407e-31, 5.22711793e-31, 5.22709756e-31], [5.22710771e-31, 5.22710360e-31, 5.22708275e-31], ..., [5.22537477e-31, 5.22539494e-31, 5.22540402e-31], [5.22541116e-31, 5.22543096e-31, 5.22543906e-31], [5.22546030e-31, 5.22549348e-31, 5.22549852e-31]], [[5.22713109e-31, 5.22712813e-31, 5.22710770e-31], [5.22712149e-31, 5.22711879e-31, 5.22709994e-31], [5.22710737e-31, 5.22710345e-31, 5.22708519e-31], ..., [5.22538468e-31, 5.22540543e-31, 5.22541645e-31], [5.22543229e-31, 5.22545842e-31, 5.22546797e-31], [5.22545942e-31, 5.22550059e-31, 5.22551111e-31]]]) - kappa(time, lev, ncol)float640.1087 0.1087 ... 0.1086 0.1086
- long_name :
- Mass extinction cross section per molecule
- units :
- $kg\ m^{-3}$
array([[[0.10868007, 0.10868007, 0.10867971], [0.10867986, 0.10867989, 0.10867952], [0.10867956, 0.10867956, 0.10867922], ..., [0.10864507, 0.1086454 , 0.10864563], [0.1086465 , 0.10864738, 0.10864763], [0.10864632, 0.1086473 , 0.10864759]], [[0.10868008, 0.10868009, 0.10867973], [0.1086799 , 0.1086799 , 0.10867953], [0.10867955, 0.10867959, 0.10867925], ..., [0.10864523, 0.10864531, 0.10864554], [0.10864694, 0.10864784, 0.10864804], [0.10864647, 0.10864764, 0.10864793]], [[0.1086801 , 0.10868013, 0.10867975], [0.10867992, 0.10867994, 0.10867957], [0.10867958, 0.10867962, 0.10867927], ..., ... ..., [0.10864348, 0.10864398, 0.10864419], [0.10864403, 0.10864427, 0.10864447], [0.10864527, 0.10864593, 0.10864614]], [[0.10868011, 0.10868 , 0.10867958], [0.10867993, 0.1086798 , 0.10867937], [0.1086796 , 0.1086795 , 0.10867907], ..., [0.10864356, 0.10864398, 0.10864418], [0.10864432, 0.10864472, 0.1086449 ], [0.10864533, 0.10864603, 0.10864613]], [[0.10868008, 0.10868001, 0.10867959], [0.10867987, 0.10867982, 0.10867944], [0.10867958, 0.1086795 , 0.10867913], ..., [0.10864376, 0.1086442 , 0.10864443], [0.10864476, 0.1086453 , 0.1086455 ], [0.10864533, 0.10864617, 0.10864639]]]) - N_s(time, lev, ncol)float642.802e+25 2.777e+25 ... 3.383e+21
- long_name :
- Number density profile
- units :
- $m^{-3}$
array([[[2.80212009e+25, 2.77707699e+25, 2.77187074e+25], [2.79273927e+25, 2.76770281e+25, 2.76161275e+25], [2.77772003e+25, 2.75280492e+25, 2.74587181e+25], ..., [7.53490060e+21, 7.49749374e+21, 7.47013248e+21], [4.98564971e+21, 4.91348959e+21, 4.89324942e+21], [3.38776361e+21, 3.33270042e+21, 3.31534923e+21]], [[2.80201992e+25, 2.77712628e+25, 2.77243115e+25], [2.79260399e+25, 2.76776772e+25, 2.76208376e+25], [2.77753059e+25, 2.75291750e+25, 2.74627162e+25], ..., [7.51914114e+21, 7.50550097e+21, 7.48177419e+21], [4.95094211e+21, 4.87630470e+21, 4.85847282e+21], [3.38032760e+21, 3.31377557e+21, 3.29650082e+21]], [[2.80192729e+25, 2.77741370e+25, 2.77301042e+25], [2.79247104e+25, 2.76810128e+25, 2.76265766e+25], [2.77734537e+25, 2.75324005e+25, 2.74684420e+25], ..., ... ..., [7.72711303e+21, 7.66987850e+21, 7.64255988e+21], [5.18998749e+21, 5.16829286e+21, 5.15277822e+21], [3.44673514e+21, 3.40832956e+21, 3.39736575e+21]], [[2.70842569e+25, 2.74533338e+25, 2.76275810e+25], [2.69922434e+25, 2.73614132e+25, 2.75304448e+25], [2.68449626e+25, 2.72140276e+25, 2.73721405e+25], ..., [7.72156293e+21, 7.67013560e+21, 7.64420231e+21], [5.16541946e+21, 5.13203525e+21, 5.11875478e+21], [3.44237895e+21, 3.40333325e+21, 3.39725579e+21]], [[2.70839694e+25, 2.73645787e+25, 2.75122090e+25], [2.69920879e+25, 2.72728381e+25, 2.74160666e+25], [2.68450964e+25, 2.71254190e+25, 2.72599750e+25], ..., [7.69405158e+21, 7.63943579e+21, 7.61434664e+21], [5.12931285e+21, 5.08403919e+21, 5.06949506e+21], [3.44224084e+21, 3.39594057e+21, 3.38325903e+21]]]) - n_s(time, lev, ncol)float641.0 1.0 1.0 1.0 ... 1.0 1.0 1.0 1.0
- long_name :
- Refractive index
- units :
- 1
array([[[1.00030607, 1.00030333, 1.00030276], [1.00030504, 1.00030231, 1.00030164], [1.0003034 , 1.00030068, 1.00029992], ..., [1.00000008, 1.00000008, 1.00000008], [1.00000005, 1.00000005, 1.00000005], [1.00000004, 1.00000004, 1.00000004]], [[1.00030606, 1.00030334, 1.00030282], [1.00030503, 1.00030231, 1.00030169], [1.00030338, 1.00030069, 1.00029997], ..., [1.00000008, 1.00000008, 1.00000008], [1.00000005, 1.00000005, 1.00000005], [1.00000004, 1.00000004, 1.00000004]], [[1.00030605, 1.00030337, 1.00030289], [1.00030501, 1.00030235, 1.00030176], [1.00030336, 1.00030073, 1.00030003], ..., ... ..., [1.00000008, 1.00000008, 1.00000008], [1.00000006, 1.00000006, 1.00000006], [1.00000004, 1.00000004, 1.00000004]], [[1.00029583, 1.00029986, 1.00030177], [1.00029483, 1.00029886, 1.00030071], [1.00029322, 1.00029725, 1.00029898], ..., [1.00000008, 1.00000008, 1.00000008], [1.00000006, 1.00000006, 1.00000006], [1.00000004, 1.00000004, 1.00000004]], [[1.00029583, 1.0002989 , 1.00030051], [1.00029483, 1.00029789, 1.00029946], [1.00029322, 1.00029628, 1.00029775], ..., [1.00000008, 1.00000008, 1.00000008], [1.00000006, 1.00000006, 1.00000006], [1.00000004, 1.00000004, 1.00000004]]]) - sub_col_beta_p_tot_strat(subcolumn, time, lev, ncol)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Backscatter coefficient from all stratiform hydrometeors
- units :
- $m^{-1} sr^{-1}$
- Processing method :
- Bulk (radiation logic)
- Ice scattering database :
- m-D_A-D (D. Mitchell)
array([[[[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., ... ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]]]]) - sub_col_alpha_p_tot_strat(subcolumn, time, lev, ncol)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Extinction coefficient from all stratiform hydrometeors
- units :
- $m^{-1}$
- Processing method :
- Bulk (radiation logic)
- Ice scattering database :
- m-D_A-D (D. Mitchell)
array([[[[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., ... ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]]]]) - sub_col_OD_tot_strat(subcolumn, time, lev, ncol)float64nan nan nan ... 30.94 20.9 20.75
- long_name :
- Cumulative optical depth at model layer base from all stratiform hydrometeors
- units :
- 1
- Processing method :
- Bulk (radiation logic)
- Ice scattering database :
- m-D_A-D (D. Mitchell)
array([[[[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., [17.14330429, 20.52316798, 15.65262676], [17.14330429, 20.52316798, 15.65262676], [17.14330429, 20.52316798, 15.65262676]], [[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., [15.83054956, 19.56877173, 16.48598632], [15.83054956, 19.56877173, 16.48598632], [15.83054956, 19.56877173, 16.48598632]], [[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., ... ..., [23.42118876, 15.4967975 , 8.35784375], [23.42118876, 15.4967975 , 8.35784375], [23.42118876, 15.4967975 , 8.35784375]], [[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., [31.84092996, 21.83319126, 16.71047916], [31.84092996, 21.83319126, 16.71047916], [31.84092996, 21.83319126, 16.71047916]], [[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., [30.94317789, 20.90443781, 20.7459365 ], [30.94317789, 20.90443781, 20.7459365 ], [30.94317789, 20.90443781, 20.7459365 ]]]]) - sub_col_beta_p_cl_strat(subcolumn, time, lev, ncol)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Particulate backscatter cross section from stratiform cl hydrometeors
- units :
- $m^{-1} sr^{-1}$
- Processing method :
- Bulk (radiation logic)
- Ice scattering database :
- m-D_A-D (D. Mitchell)
array([[[[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., ... ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]]]]) - sub_col_alpha_p_cl_strat(subcolumn, time, lev, ncol)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Particulate extinction cross section from stratiform cl hydrometeors
- units :
- $m^{-1}$
- Processing method :
- Bulk (radiation logic)
- Ice scattering database :
- m-D_A-D (D. Mitchell)
array([[[[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., ... ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]]]]) - sub_col_OD_cl_strat(subcolumn, time, lev, ncol)float64nan nan nan ... 23.49 6.234 8.599
- long_name :
- Cumulative optical depth at model layer base from stratiform cl hydrometeors
- units :
- 1
- Processing method :
- Bulk (radiation logic)
- Ice scattering database :
- m-D_A-D (D. Mitchell)
array([[[[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., [17.14330429, 20.42890334, 15.26308484], [17.14330429, 20.42890334, 15.26308484], [17.14330429, 20.42890334, 15.26308484]], [[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., [15.83054956, 19.51257512, 16.30535001], [15.83054956, 19.51257512, 16.30535001], [15.83054956, 19.51257512, 16.30535001]], [[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., ... ..., [12.72656987, 3.62611793, nan], [12.72656987, 3.62611793, nan], [12.72656987, 3.62611793, nan]], [[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., [22.31658271, 7.25559343, 7.71856522], [22.31658271, 7.25559343, 7.71856522], [22.31658271, 7.25559343, 7.71856522]], [[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., [23.48838799, 6.23429879, 8.59854985], [23.48838799, 6.23429879, 8.59854985], [23.48838799, 6.23429879, 8.59854985]]]]) - sub_col_alpha_p_ci_strat(subcolumn, time, lev, ncol)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Particulate extinction cross section from stratiform ci hydrometeors
- units :
- $m^{-1}$
- Processing method :
- Bulk (radiation logic)
- Ice scattering database :
- m-D_A-D (D. Mitchell)
array([[[[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., ... ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]]]]) - sub_col_beta_p_ci_strat(subcolumn, time, lev, ncol)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Particulate backscatter cross section from stratiform ci hydrometeors
- units :
- $m^{-1} sr^{-1}$
- Processing method :
- Bulk (radiation logic)
- Ice scattering database :
- m-D_A-D (D. Mitchell)
array([[[[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., ... ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]]]]) - sub_col_OD_ci_strat(subcolumn, time, lev, ncol)float64nan nan nan ... 0.8173 0.6555
- long_name :
- Cumulative optical depth at model layer base from stratiform ci hydrometeors
- units :
- 1
- Processing method :
- Bulk (radiation logic)
- Ice scattering database :
- m-D_A-D (D. Mitchell)
array([[[[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., [ nan, 0.02802345, 0.12366886], [ nan, 0.02802345, 0.12366886], [ nan, 0.02802345, 0.12366886]], [[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., [ nan, nan, 0.1014499 ], [ nan, nan, 0.1014499 ], [ nan, nan, 0.1014499 ]], [[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., ... ..., [0.51675301, 0.7197492 , 0.67561573], [0.51675301, 0.7197492 , 0.67561573], [0.51675301, 0.7197492 , 0.67561573]], [[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., [0.42066257, 0.99352207, 0.76734315], [0.42066257, 0.99352207, 0.76734315], [0.42066257, 0.99352207, 0.76734315]], [[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., [0.48372899, 0.81730486, 0.65546422], [0.48372899, 0.81730486, 0.65546422], [0.48372899, 0.81730486, 0.65546422]]]]) - sub_col_alpha_p_pi_strat(subcolumn, time, lev, ncol)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Particulate extinction cross section from stratiform pi hydrometeors
- units :
- $m^{-1}$
- Processing method :
- Bulk (radiation logic)
- Ice scattering database :
- m-D_A-D (D. Mitchell)
array([[[[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., ... ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]]]]) - sub_col_beta_p_pi_strat(subcolumn, time, lev, ncol)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Particulate backscatter cross section from stratiform pi hydrometeors
- units :
- $m^{-1} sr^{-1}$
- Processing method :
- Bulk (radiation logic)
- Ice scattering database :
- m-D_A-D (D. Mitchell)
array([[[[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., ... ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]]]]) - sub_col_OD_pi_strat(subcolumn, time, lev, ncol)float64nan nan nan ... 6.971 13.85 11.49
- long_name :
- Cumulative optical depth at model layer base from stratiform pi hydrometeors
- units :
- 1
- Processing method :
- Bulk (radiation logic)
- Ice scattering database :
- m-D_A-D (D. Mitchell)
array([[[[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., [ nan, 6.62411921e-02, 2.65873059e-01], [ nan, 6.62411921e-02, 2.65873059e-01], [ nan, 6.62411921e-02, 2.65873059e-01]], [[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., [ nan, 5.61966131e-02, 7.91864087e-02], [ nan, 5.61966131e-02, 7.91864087e-02], [ nan, 5.61966131e-02, 7.91864087e-02]], [[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., ... ..., [1.01778659e+01, 1.11509304e+01, 7.68222802e+00], [1.01778659e+01, 1.11509304e+01, 7.68222802e+00], [1.01778659e+01, 1.11509304e+01, 7.68222802e+00]], [[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., [9.10368468e+00, 1.35840758e+01, 8.22457079e+00], [9.10368468e+00, 1.35840758e+01, 8.22457079e+00], [9.10368468e+00, 1.35840758e+01, 8.22457079e+00]], [[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., [6.97106091e+00, 1.38528342e+01, 1.14919224e+01], [6.97106091e+00, 1.38528342e+01, 1.14919224e+01], [6.97106091e+00, 1.38528342e+01, 1.14919224e+01]]]]) - sub_col_beta_att_tot_strat(subcolumn, time, lev, ncol)float641.718e-06 1.703e-06 ... 1.582e-28
- long_name :
- Total attenuated backscatter from all stratiform hydrometeors (including atmospheric extinction)
- units :
- $m^{-1} sr^{-1}$
- Processing method :
- Bulk (radiation logic)
- Ice scattering database :
- m-D_A-D (D. Mitchell)
array([[[[1.71796754e-06, 1.70261388e-06, 1.69941618e-06], [1.71034996e-06, 1.69502508e-06, 1.69130215e-06], [1.69819484e-06, 1.68298289e-06, 1.67877241e-06], ..., [4.72604512e-25, 5.45797034e-28, 9.26045754e-24], [3.12708316e-25, 3.57688191e-28, 6.06598102e-24], [2.12483080e-25, 2.42607838e-28, 4.10986179e-24]], [[1.71790628e-06, 1.70264421e-06, 1.69976027e-06], [1.71026734e-06, 1.69506402e-06, 1.69158977e-06], [1.69807750e-06, 1.68304943e-06, 1.67901475e-06], ..., [6.51333615e-24, 3.68488212e-27, 1.75149187e-24], [4.28865891e-24, 2.39406740e-27, 1.13737815e-24], [2.92809336e-24, 1.62690488e-27, 7.71706571e-25]], [[1.71784986e-06, 1.70282131e-06, 1.70011561e-06], [1.71018571e-06, 1.69526795e-06, 1.69194065e-06], [1.69796321e-06, 1.68324430e-06, 1.67936201e-06], ..., ... ..., [1.71349726e-30, 1.29777903e-23, 2.05551294e-17], [1.15087003e-30, 8.74484544e-24, 1.38584772e-17], [7.64305855e-31, 5.76696806e-24, 9.13728847e-18]], [[1.66052471e-06, 1.68315087e-06, 1.69382716e-06], [1.65310539e-06, 1.67570523e-06, 1.68605912e-06], [1.64126915e-06, 1.66381204e-06, 1.67349429e-06], ..., [8.32451509e-38, 4.07041322e-29, 1.14322480e-24], [5.56870170e-38, 2.72345178e-29, 7.65523113e-25], [3.71112374e-38, 1.80606777e-29, 5.08067580e-25]], [[1.66050650e-06, 1.67770958e-06, 1.68675414e-06], [1.65309593e-06, 1.67028314e-06, 1.67905693e-06], [1.64127956e-06, 1.65840068e-06, 1.66664265e-06], ..., [4.99615106e-37, 2.59856044e-28, 3.55953744e-28], [3.33069770e-37, 1.72932426e-28, 2.36985356e-28], [2.23518695e-37, 1.15511559e-28, 1.58157560e-28]]]]) - sub_col_beta_p_tot(subcolumn, time, lev, ncol)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Total backscatter coefficient (convective + stratiform)
- units :
- $m^{-1} sr^{-1}$
- Processing method :
- Bulk (radiation logic)
- Ice scattering database :
- m-D_A-D (D. Mitchell)
array([[[[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., ... ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]]]]) - sub_col_alpha_p_tot(subcolumn, time, lev, ncol)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Total extinction coefficient (convective + stratiform)
- units :
- $m^{-1}$
- Processing method :
- Bulk (radiation logic)
- Ice scattering database :
- m-D_A-D (D. Mitchell)
array([[[[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., ... ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]]]]) - sub_col_OD_tot(subcolumn, time, lev, ncol)float64nan nan nan ... 30.94 20.9 20.75
- long_name :
- Total cumulative optical depth at layer base (convective + stratiform)
- units :
- 1
- Processing method :
- Bulk (radiation logic)
- Ice scattering database :
- m-D_A-D (D. Mitchell)
array([[[[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., [17.14330429, 20.52316798, 15.65262676], [17.14330429, 20.52316798, 15.65262676], [17.14330429, 20.52316798, 15.65262676]], [[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., [15.83054956, 19.56877173, 16.48598632], [15.83054956, 19.56877173, 16.48598632], [15.83054956, 19.56877173, 16.48598632]], [[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., ... ..., [23.42118876, 15.4967975 , 8.35784375], [23.42118876, 15.4967975 , 8.35784375], [23.42118876, 15.4967975 , 8.35784375]], [[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., [31.84092996, 21.83319126, 16.71047916], [31.84092996, 21.83319126, 16.71047916], [31.84092996, 21.83319126, 16.71047916]], [[ nan, nan, nan], [ nan, nan, nan], [ nan, nan, nan], ..., [30.94317789, 20.90443781, 20.7459365 ], [30.94317789, 20.90443781, 20.7459365 ], [30.94317789, 20.90443781, 20.7459365 ]]]]) - sub_col_beta_att_tot(subcolumn, time, lev, ncol)float641.718e-06 1.703e-06 ... 1.582e-28
- long_name :
- Total attenuated backscatter coefficient (convective + stratiform)
- units :
- $m^{-1} sr^{-1}$
array([[[[1.71796754e-06, 1.70261388e-06, 1.69941618e-06], [1.71034996e-06, 1.69502508e-06, 1.69130215e-06], [1.69819484e-06, 1.68298289e-06, 1.67877241e-06], ..., [4.72604512e-25, 5.45797034e-28, 9.26045754e-24], [3.12708316e-25, 3.57688191e-28, 6.06598102e-24], [2.12483080e-25, 2.42607838e-28, 4.10986179e-24]], [[1.71790628e-06, 1.70264421e-06, 1.69976027e-06], [1.71026734e-06, 1.69506402e-06, 1.69158977e-06], [1.69807750e-06, 1.68304943e-06, 1.67901475e-06], ..., [6.51333615e-24, 3.68488212e-27, 1.75149187e-24], [4.28865891e-24, 2.39406740e-27, 1.13737815e-24], [2.92809336e-24, 1.62690488e-27, 7.71706571e-25]], [[1.71784986e-06, 1.70282131e-06, 1.70011561e-06], [1.71018571e-06, 1.69526795e-06, 1.69194065e-06], [1.69796321e-06, 1.68324430e-06, 1.67936201e-06], ..., ... ..., [1.71349726e-30, 1.29777903e-23, 2.05551294e-17], [1.15087003e-30, 8.74484544e-24, 1.38584772e-17], [7.64305855e-31, 5.76696806e-24, 9.13728847e-18]], [[1.66052471e-06, 1.68315087e-06, 1.69382716e-06], [1.65310539e-06, 1.67570523e-06, 1.68605912e-06], [1.64126915e-06, 1.66381204e-06, 1.67349429e-06], ..., [8.32451509e-38, 4.07041322e-29, 1.14322480e-24], [5.56870170e-38, 2.72345178e-29, 7.65523113e-25], [3.71112374e-38, 1.80606777e-29, 5.08067580e-25]], [[1.66050650e-06, 1.67770958e-06, 1.68675414e-06], [1.65309593e-06, 1.67028314e-06, 1.67905693e-06], [1.64127956e-06, 1.65840068e-06, 1.66664265e-06], ..., [4.99615106e-37, 2.59856044e-28, 3.55953744e-28], [3.33069770e-37, 1.72932426e-28, 2.36985356e-28], [2.23518695e-37, 1.15511559e-28, 1.58157560e-28]]]]) - sub_col_LDR_strat(subcolumn, time, lev, ncol)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Linear depolarization ratio in strat
- units :
- 1
array([[[[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., ... ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]]]]) - sub_col_LDR_tot(subcolumn, time, lev, ncol)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Linear depolarization ratio (convective + stratiform)
- units :
- 1
array([[[[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., ... ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]]]]) - ext_mask(subcolumn, time, lev, ncol)float640.0 0.0 0.0 0.0 ... 2.0 2.0 2.0 2.0
- long_name :
- Extinction mask at layer base based on optical thickness considerations (convective + stratiform; calculated from surface)
- units :
- 2 = Signal extinct, 1 = layer where signal becomes extinct, 0 = signal not extinct
array([[[[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [2., 2., 2.], [2., 2., 2.], [2., 2., 2.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [2., 2., 2.], [2., 2., 2.], [2., 2., 2.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., ... ..., [2., 2., 2.], [2., 2., 2.], [2., 2., 2.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [2., 2., 2.], [2., 2., 2.], [2., 2., 2.]], [[0., 0., 0.], [0., 0., 0.], [0., 0., 0.], ..., [2., 2., 2.], [2., 2., 2.], [2., 2., 2.]]]]) - phase_mask_HSRL_all_hyd(subcolumn, time, lev, ncol)float64nan nan nan nan ... nan nan nan nan
- long_name :
- HSRL phase classification mask (convective + stratiform)
- units :
- Unitless
- legend :
- ['liquid', 'ice', 'undef1', 'undef2']
array([[[[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., ... ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]]]]) - strat_COSP_phase_mask(subcolumn, time, lev, ncol)float64nan nan nan nan ... nan nan nan nan
- long_name :
- COSP emulation phase classification mask (strat)
- units :
- Unitless
- legend :
- ['liquid', 'ice', 'undef']
array([[[[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., ... ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]]]]) - COSP_phase_mask_all_hyd(subcolumn, time, lev, ncol)float64nan nan nan nan ... nan nan nan nan
- long_name :
- COSP emulation phase classification mask (convective + stratiform)
- units :
- Unitless
- legend :
- ['liquid', 'ice', 'undef']
array([[[[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., ... ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]]]]) - kappa_o2(time, lev, ncol)float640.01881 0.0184 ... 5.519e-10
- long_name :
- Gaseous attenuation due to O2
- units :
- $dB\ km^{-1}$
array([[[1.88135504e-02, 1.84022439e-02, 1.84128876e-02], [1.87101169e-02, 1.82996260e-02, 1.82931790e-02], [1.85448194e-02, 1.81376740e-02, 1.81147798e-02], ..., [2.78740387e-09, 2.74814682e-09, 2.71966008e-09], [1.19642799e-09, 1.14773350e-09, 1.13431017e-09], [5.53965072e-10, 5.28687870e-10, 5.20881011e-10]], [[1.88068383e-02, 1.83955209e-02, 1.84150661e-02], [1.87027717e-02, 1.82932360e-02, 1.82936937e-02], [1.85364874e-02, 1.81322440e-02, 1.81139999e-02], ..., [2.77082151e-09, 2.75652037e-09, 2.73175789e-09], [1.17284270e-09, 1.12315143e-09, 1.11148545e-09], [5.50506592e-10, 5.20176478e-10, 5.12485507e-10]], [[1.88003019e-02, 1.83924426e-02, 1.84167734e-02], [1.86954943e-02, 1.82910543e-02, 1.82953326e-02], [1.85282724e-02, 1.81299150e-02, 1.81156642e-02], ..., ... ..., [2.99487120e-09, 2.93208262e-09, 2.90241671e-09], [1.34153933e-09, 1.32561888e-09, 1.31430971e-09], [5.81892325e-10, 5.63603224e-10, 5.58451656e-10]], [[1.72922257e-02, 1.79044536e-02, 1.82981475e-02], [1.71947197e-02, 1.78059968e-02, 1.81888018e-02], [1.70388930e-02, 1.76481871e-02, 1.80088022e-02], ..., [2.98874519e-09, 2.93236294e-09, 2.90419452e-09], [1.32351974e-09, 1.29928651e-09, 1.28972717e-09], [5.79798687e-10, 5.61251892e-10, 5.58400012e-10]], [[1.72988054e-02, 1.77601791e-02, 1.80994001e-02], [1.72015014e-02, 1.76623047e-02, 1.79923417e-02], [1.70461387e-02, 1.75048163e-02, 1.78171776e-02], ..., [2.95849637e-09, 2.89903578e-09, 2.87198445e-09], [1.29732299e-09, 1.26495403e-09, 1.25466833e-09], [5.79732283e-10, 5.57784244e-10, 5.51868324e-10]]]) - kappa_wv(time, lev, ncol)float641.2e-06 1.2e-06 ... 1.2e-06 1.2e-06
- long_name :
- Gaseous attenuation due to water vapor
- units :
- $dB\ km^{-1}$
array([[[1.2e-06, 1.2e-06, 1.2e-06], [1.2e-06, 1.2e-06, 1.2e-06], [1.2e-06, 1.2e-06, 1.2e-06], ..., [1.2e-06, 1.2e-06, 1.2e-06], [1.2e-06, 1.2e-06, 1.2e-06], [1.2e-06, 1.2e-06, 1.2e-06]], [[1.2e-06, 1.2e-06, 1.2e-06], [1.2e-06, 1.2e-06, 1.2e-06], [1.2e-06, 1.2e-06, 1.2e-06], ..., [1.2e-06, 1.2e-06, 1.2e-06], [1.2e-06, 1.2e-06, 1.2e-06], [1.2e-06, 1.2e-06, 1.2e-06]], [[1.2e-06, 1.2e-06, 1.2e-06], [1.2e-06, 1.2e-06, 1.2e-06], [1.2e-06, 1.2e-06, 1.2e-06], ..., ... ..., [1.2e-06, 1.2e-06, 1.2e-06], [1.2e-06, 1.2e-06, 1.2e-06], [1.2e-06, 1.2e-06, 1.2e-06]], [[1.2e-06, 1.2e-06, 1.2e-06], [1.2e-06, 1.2e-06, 1.2e-06], [1.2e-06, 1.2e-06, 1.2e-06], ..., [1.2e-06, 1.2e-06, 1.2e-06], [1.2e-06, 1.2e-06, 1.2e-06], [1.2e-06, 1.2e-06, 1.2e-06]], [[1.2e-06, 1.2e-06, 1.2e-06], [1.2e-06, 1.2e-06, 1.2e-06], [1.2e-06, 1.2e-06, 1.2e-06], ..., [1.2e-06, 1.2e-06, 1.2e-06], [1.2e-06, 1.2e-06, 1.2e-06], [1.2e-06, 1.2e-06, 1.2e-06]]]) - kappa_att(time, lev, ncol)float640.01881 0.0184 ... 1.201e-06
- long_name :
- Gaseous attenuation due to O2 and water vapor
- units :
- $dB\ km^{-1}$
array([[[1.88147504e-02, 1.84034439e-02, 1.84140876e-02], [1.87113169e-02, 1.83008260e-02, 1.82943790e-02], [1.85460194e-02, 1.81388740e-02, 1.81159798e-02], ..., [1.20278740e-06, 1.20274815e-06, 1.20271966e-06], [1.20119643e-06, 1.20114773e-06, 1.20113431e-06], [1.20055397e-06, 1.20052869e-06, 1.20052088e-06]], [[1.88080383e-02, 1.83967209e-02, 1.84162661e-02], [1.87039717e-02, 1.82944360e-02, 1.82948937e-02], [1.85376874e-02, 1.81334440e-02, 1.81151999e-02], ..., [1.20277082e-06, 1.20275652e-06, 1.20273176e-06], [1.20117284e-06, 1.20112315e-06, 1.20111149e-06], [1.20055051e-06, 1.20052018e-06, 1.20051249e-06]], [[1.88015019e-02, 1.83936426e-02, 1.84179734e-02], [1.86966943e-02, 1.82922543e-02, 1.82965326e-02], [1.85294724e-02, 1.81311150e-02, 1.81168642e-02], ..., ... ..., [1.20299487e-06, 1.20293208e-06, 1.20290242e-06], [1.20134154e-06, 1.20132562e-06, 1.20131431e-06], [1.20058189e-06, 1.20056360e-06, 1.20055845e-06]], [[1.72934257e-02, 1.79056536e-02, 1.82993475e-02], [1.71959197e-02, 1.78071968e-02, 1.81900018e-02], [1.70400930e-02, 1.76493871e-02, 1.80100022e-02], ..., [1.20298875e-06, 1.20293236e-06, 1.20290419e-06], [1.20132352e-06, 1.20129929e-06, 1.20128973e-06], [1.20057980e-06, 1.20056125e-06, 1.20055840e-06]], [[1.73000054e-02, 1.77613791e-02, 1.81006001e-02], [1.72027014e-02, 1.76635047e-02, 1.79935417e-02], [1.70473387e-02, 1.75060163e-02, 1.78183776e-02], ..., [1.20295850e-06, 1.20289904e-06, 1.20287198e-06], [1.20129732e-06, 1.20126495e-06, 1.20125467e-06], [1.20057973e-06, 1.20055778e-06, 1.20055187e-06]]]) - sub_col_Ze_tot_strat(subcolumn, time, lev, ncol)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Equivalent radar reflectivity factor from all stratiform hydrometeors
- units :
- dBZ
- Processing method :
- Bulk (radiation logic)
- Ice scattering database :
- C6
array([[[[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., ... ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]]]]) - sub_col_Ze_cl_strat(subcolumn, time, lev, ncol)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Equivalent radar reflectivity factor from stratiform cl hydrometeors
- units :
- dBZ
- Processing method :
- Bulk (radiation logic)
- Ice scattering database :
- C6
array([[[[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., ... ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]]]]) - sub_col_Ze_ci_strat(subcolumn, time, lev, ncol)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Equivalent radar reflectivity factor from stratiform ci hydrometeors
- units :
- dBZ
- Processing method :
- Bulk (radiation logic)
- Ice scattering database :
- C6
array([[[[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., ... ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]]]]) - sub_col_Ze_pi_strat(subcolumn, time, lev, ncol)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Equivalent radar reflectivity factor from stratiform pi hydrometeors
- units :
- dBZ
- Processing method :
- Bulk (radiation logic)
- Ice scattering database :
- C6
array([[[[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., ... ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]]]]) - hyd_ext_strat(subcolumn, time, lev, ncol)float641.0 1.0 1.0 nan ... nan nan nan nan
- long_name :
- Two-way strat hydrometeor transmittance at model layer base
- units :
- 1
- Processing method :
- Bulk (radiation logic)
- Ice scattering database :
- C6
array([[[[ 1., 1., 1.], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[ 1., 1., 1.], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[ 1., 1., 1.], [nan, nan, nan], [nan, nan, nan], ..., ... ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[ 1., 1., 1.], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[ 1., 1., 1.], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]]]]) - atm_ext(time, lev, ncol)float641.0 1.0 1.0 ... 0.9605 0.96 0.9601
- long_name :
- Two-way atmospheric transmittance due to H2O and O2 at model layer base
- units :
- 1
array([[[1. , 1. , 1. ], [0.99967752, 0.99968303, 0.99968438], [0.9991638 , 0.99917819, 0.9991822 ], ..., [0.95813723, 0.9582352 , 0.95867942], [0.95813566, 0.95823361, 0.95867783], [0.95813408, 0.95823201, 0.95867621]], [[1. , 1. , 1. ], [0.99967753, 0.99968298, 0.99968423], [0.99916381, 0.99917807, 0.99918182], ..., [0.95815047, 0.95823242, 0.95867129], [0.9581489 , 0.95823083, 0.95866969], [0.95814731, 0.95822921, 0.95866806]], [[1. , 1. , 1. ], [0.99967753, 0.99968289, 0.99968407], [0.99916383, 0.9991778 , 0.9991814 ], ..., ... ..., [0.96040543, 0.95956562, 0.9597803 ], [0.96040391, 0.95956409, 0.95977876], [0.96040237, 0.95956254, 0.9597772 ]], [[1. , 1. , 1. ], [0.99969765, 0.99968987, 0.99968648], [0.99921636, 0.99919606, 0.99918748], ..., [0.96046855, 0.95977018, 0.95995982], [0.96046703, 0.95976864, 0.95995828], [0.96046548, 0.95976708, 0.95995672]], [[1. , 1. , 1. ], [0.99969769, 0.99969177, 0.99968893], [0.99921645, 0.99920099, 0.99919384], ..., [0.96048459, 0.9599639 , 0.96014071], [0.96048306, 0.95996235, 0.96013916], [0.96048151, 0.95996078, 0.96013758]]]) - sub_col_Ze_att_tot_strat(subcolumn, time, lev, ncol)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Attenuated equivalent radar reflectivity factor from all stratiform hydrometeors
- units :
- dBZ
- Processing method :
- Bulk (radiation logic)
- Ice scattering database :
- C6
array([[[[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., ... ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]]]]) - Ze_min(time, lev, ncol)float32-87.04 -78.36 ... -12.82 -12.81
- long_name :
- Minimum discernable radar reflectivity factor
- units :
- dBZ
array([[[-87.042336 , -78.35667 , -69.4135 ], [-74.68699 , -71.65338 , -66.4074 ], [-67.775055 , -66.25527 , -63.068615 ], ..., [-13.675034 , -13.653564 , -13.645561 ], [-13.221008 , -13.196266 , -13.186928 ], [-12.785339 , -12.75518 , -12.744308 ]], [[-87.039444 , -78.355064 , -69.41309 ], [-74.68403 , -71.650314 , -66.40615 ], [-67.77194 , -66.25157 , -63.066578 ], ..., [-13.680695 , -13.657928 , -13.64904 ], [-13.224342 , -13.198906 , -13.188942 ], [-12.786545 , -12.754829 , -12.743439 ]], [[-87.03662 , -78.35356 , -69.412636 ], [-74.68112 , -71.64747 , -66.4048 ], [-67.76888 , -66.24811 , -63.064453 ], ..., ... ..., [-13.73703 , -13.708054 , -13.6944275], [-13.294189 , -13.264137 , -13.2497635], [-12.867027 , -12.83519 , -12.820213 ]], [[-86.870895 , -78.338745 , -69.41385 ], [-74.514824 , -71.61903 , -66.40872 ], [-67.60581 , -66.21563 , -63.07126 ], ..., [-13.735176 , -13.705154 , -13.691727 ], [-13.2912445, -13.259865 , -13.245804 ], [-12.862976 , -12.829361 , -12.815063 ]], [[-86.8751 , -78.332405 , -69.410324 ], [-74.51906 , -71.60687 , -66.39839 ], [-67.6102 , -66.20136 , -63.055935 ], ..., [-13.736176 , -13.704041 , -13.689941 ], [-13.289894 , -13.255875 , -13.24115 ], [-12.8602295, -12.823151 , -12.807739 ]]], dtype=float32) - sub_col_Ze_tot(subcolumn, time, lev, ncol)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Total (convective + stratiform) equivalent radar reflectivity factor
- units :
- dBZ
array([[[[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., ... ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]]]]) - sub_col_Ze_att_tot(subcolumn, time, lev, ncol)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Total (convective + stratiform) attenuated (hydrometeor + gaseous) equivalent radar reflectivity factor
- units :
- dBZ
array([[[[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., ... ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]]]]) - detect_mask(subcolumn, time, lev, ncol)boolFalse False False ... False False
- long_name :
- Radar detectability mask
- units :
- 1 = radar signal below noise floor, 0 = signal detected
array([[[[False, False, False], [False, False, False], [False, False, False], ..., [False, False, False], [False, False, False], [False, False, False]], [[False, False, False], [False, False, False], [False, False, False], ..., [False, False, False], [False, False, False], [False, False, False]], [[False, False, False], [False, False, False], [False, False, False], ..., ... ..., [False, False, False], [False, False, False], [False, False, False]], [[False, False, False], [False, False, False], [False, False, False], ..., [False, False, False], [False, False, False], [False, False, False]], [[False, False, False], [False, False, False], [False, False, False], ..., [False, False, False], [False, False, False], [False, False, False]]]]) - phase_mask_KAZR_sounding_all_hyd(subcolumn, time, lev, ncol)float64nan nan nan nan ... nan nan nan nan
- long_name :
- KAZR-sounding cloud and precipitation detection output (convective + stratiform)
- units :
- Unitless
- legend :
- ['Cloud', 'Mixed', 'Precip']
array([[[[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., ... ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]], [[nan, nan, nan], [nan, nan, nan], [nan, nan, nan], ..., [nan, nan, nan], [nan, nan, nan], [nan, nan, nan]]]])
- Conventions :
- CF-1.0
- source :
- CAM
- case :
- EAMv1_ARM_Freerun_1
- title :
- UNSET
- logname :
- yshi
- host :
- cori07
- Version :
- $Name$
- revision_Id :
- $Id$
- initial_file :
- /global/cfs/cdirs/e3sm/inputdata/atm/cam/inic/homme/cami_mam3_Linoz_ne30np4_L72_c160214.nc
- topography_file :
- /global/cfs/cdirs/e3sm/inputdata/atm/cam/topo/USGS-gtopo30_ne30np4_16xdel2-PFC-consistentSGH.nc
- time_period_freq :
- hour_1
- _file_dates :
- ['20121031']
- _file_times :
- ['010000']
- _datastream :
- act_datastream
- _arm_standards_flag :
- 0
Visualization#
We can plot EMC² output using matplotlib, xarray’s internal methods, or EMC²’s emc2.plotting.SubcolumnDisplay object, which is based on the ACT package. Here, using the SubcolumnDisplay’s plot_subcolumn_timeseries method, we show the height x time curtain of the first subcolumn (out of the 50 specified) in the grid cell closest to the coordinates of the Utqiagvik specified using the lat_sel and lon_sel keywords. The hatch patterns invoked by setting hatched_mask=True, designate subcolumn bins not detected by the simulated instrument.
# set Matplotlib's shading parameter
mpl.rcParams['pcolor.shading'] = 'auto'
# Set input parameters.
cmap = 'viridis'
field_to_plot = ["sub_col_beta_p_tot", "sub_col_alpha_p_tot", "sub_col_OD_tot", "sub_col_Ze_att_tot"]
vmin_max = [(1e-8,1e-3), (1e-6, 1e-1), (0, 16), (-50., 10.)]
log_plot = [True, True, False, False]
is_radar_field = [False, False, False, True]
y_range = (200., 1e3) # in hPa
subcol_ind = 0
NSA_coords = {"lat": 71.32, "lon": -156.61}
# Generate a SubcolumnDisplay object for coords closest to the NSA site
model_display = emc2.plotting.SubcolumnDisplay(my_e3sm, subplot_shape=(1, 4), figsize=(15,5),
lat_sel=NSA_coords["lat"],
lon_sel=NSA_coords["lon"], tight_layout=True)
# set intrument detectability masks for KAZR and HSRL
Mask_array_KAZR_model = model_display.model.ds["detect_mask"].isel(subcolumn=subcol_ind)
Mask_array_HSRL_model = model_display.model.ds["ext_mask"].isel(subcolumn=subcol_ind)
# Plot variables
for ii in range(4):
if is_radar_field[ii]:
Mask_array=Mask_array_KAZR_model
else:
Mask_array=Mask_array_HSRL_model
model_display.plot_subcolumn_timeseries(field_to_plot[ii], subcol_ind, log_plot=log_plot[ii], y_range=y_range,
subplot_index=(0, ii), cmap=cmap, title='',
vmin=vmin_max[ii][0], vmax=vmin_max[ii][1],
Mask_array=Mask_array, hatched_mask=True)
cropping lat dim (lat requested = 71.32)
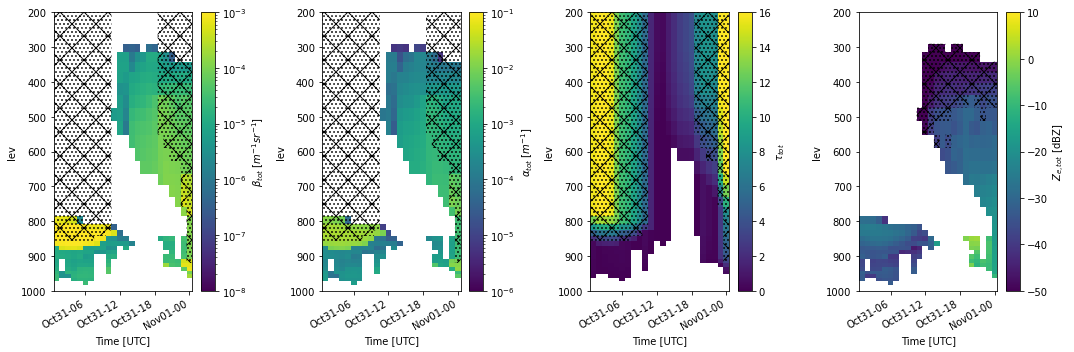
For a given subcolumn, we can also plot the output from one of the phase classification algorithms (HSRL-based classification in this case) by using the plot_subcolumn_timeseries method followed by calling the change_plot_to_class_mask.
Another option is to use the phase classification output and the emc2.simulator.classification.calculate_phase_ratio method to calcualte the hydrometeor phase partitioning based on all subcolumns, which we can then plot. Here, we also use the SubcolumnDisplay’s fig.savefig method to save the figure as a PNG file.
# Generate a SubcolumnDisplay object for coords closest to the NSA site
model_display2 = emc2.plotting.SubcolumnDisplay(my_e3sm, subplot_shape=(2, 1), figsize=(10,10),
lat_sel=NSA_coords["lat"],
lon_sel=NSA_coords["lon"], tight_layout=True)
# Plot classification mask output for a given subcolumn
_, cb2_1 = model_display2.plot_subcolumn_timeseries("phase_mask_HSRL_all_hyd", subcol_ind, y_range=y_range,
subplot_index=(0), title='')
model_display2.change_plot_to_class_mask(cb2_1, variable="phase_mask_HSRL_all_hyd", convert_zeros_to_nan=True)
# Calculate frequency phase ratio
my_e3sm = emc2.simulator.classification.calculate_phase_ratio(my_e3sm, "phase_mask_HSRL_all_hyd", [1])
# Plot frequency phase ratio based on simulated data from all subcolumns
model_display2.plot_subcolumn_timeseries("phase_mask_HSRL_all_hyd_fpr", subcol_ind, y_range=y_range,
cmap="bwr", subplot_index=(1),
cbar_label='Frequency pr (reddish - more liquid)')
model_display2.fig.savefig('HSRL_simulated_class.png', dpi=150)
cropping lat dim (lat requested = 71.32)
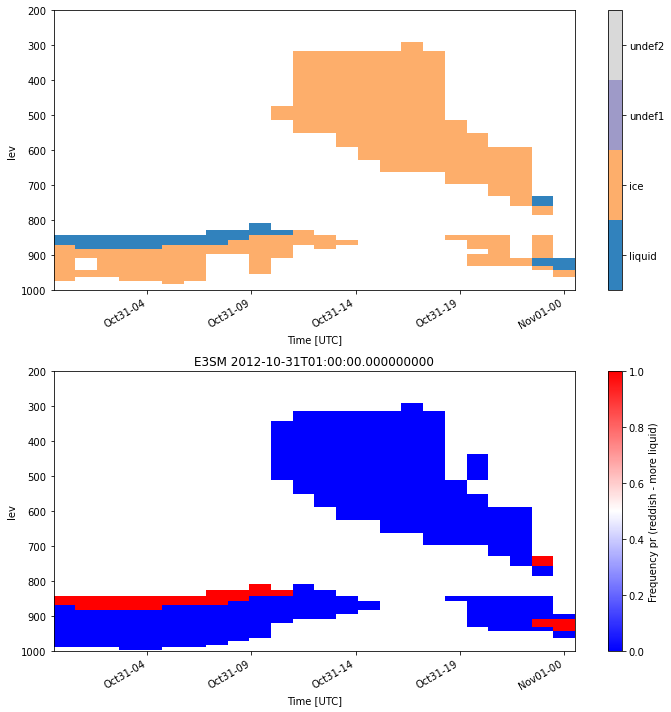
Recalling that EMC² outputs radar and lidar observables per hydrometeor class, we can examine a breakdown of hydrometeor contribution to every simulated observable. This type of analysis also enables the evaluation of the classification algorithm, generally speaking. Note that because we examine variables per hydrometeor class, we ignore the detectability constraints here. Also note that the user can quite easily edit the “cosmetics” (fontsize, etc.) when calling the plotting method, but we skip these options in this example.
# Set additional input parameters.
field_to_plot_per_class = \
["sub_col_beta_p_%s_strat", "sub_col_alpha_p_%s_strat", "sub_col_OD_%s_strat", "sub_col_Ze_%s_strat"]
hyd_class = my_e3sm.hyd_types
# generate a SubcolumnDisplay object for coords closest to the NSA site.
model_display3 = emc2.plotting.SubcolumnDisplay(my_e3sm, subplot_shape=(len(hyd_class), 4), figsize=(15, 5 * len(hyd_class)),
lat_sel=NSA_coords["lat"],
lon_sel=NSA_coords["lon"], tight_layout=True)
# Plot variables per hydrometeor class
for ii in range(4):
for class_i in range(len(hyd_class)):
if is_radar_field[ii]:
Mask_array=Mask_array_KAZR_model
else:
Mask_array=Mask_array_HSRL_model
model_display3.plot_subcolumn_timeseries(field_to_plot_per_class[ii] % hyd_class[class_i], subcol_ind,
log_plot=log_plot[ii], y_range=y_range,
subplot_index=(class_i, ii), cmap=cmap, title=hyd_class[class_i],
vmin=vmin_max[ii][0], vmax=vmin_max[ii][1])
cropping lat dim (lat requested = 71.32)
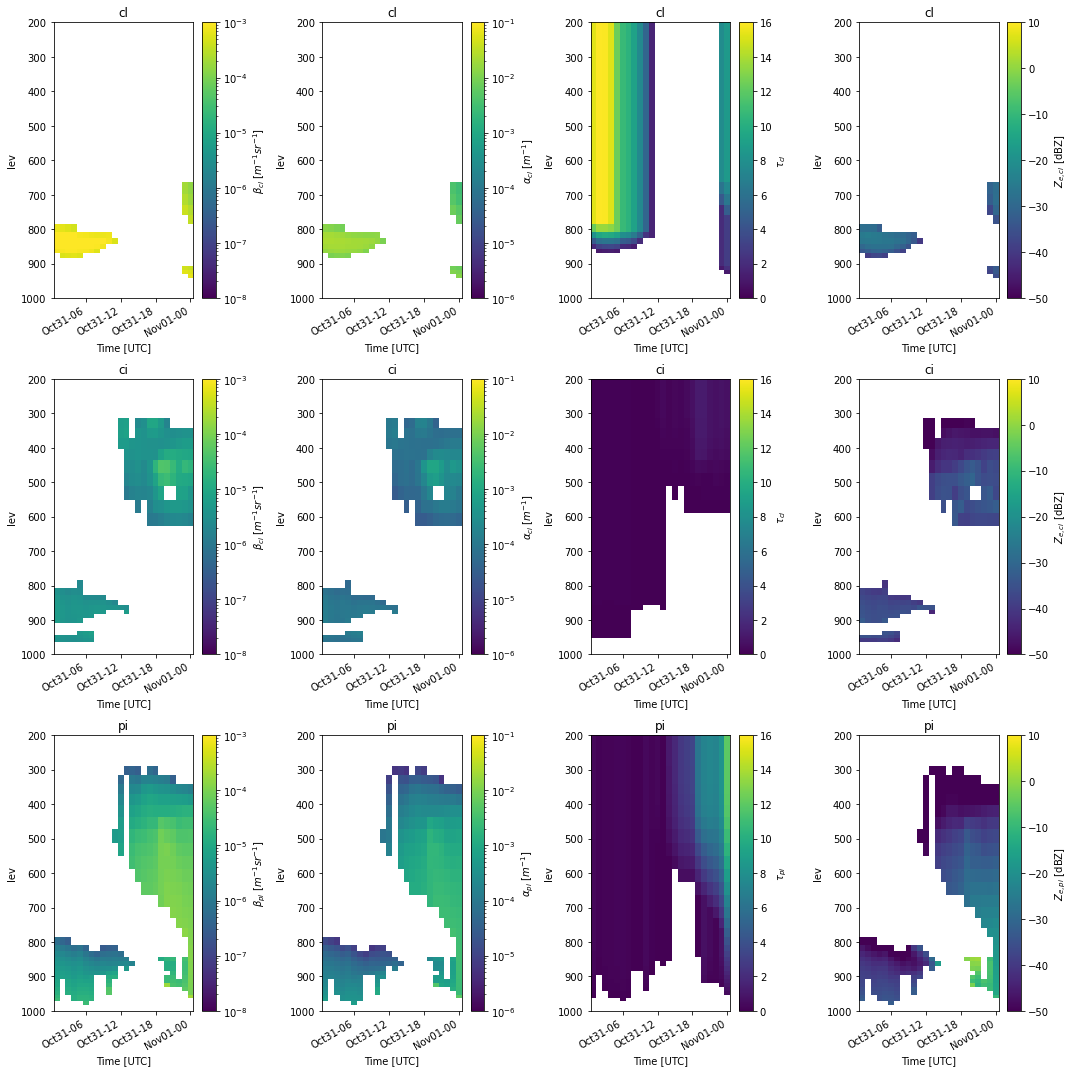
In addition to plotting the temporal evolution of a specific subcolun, we can also plot all subcolumns at a single time step using the SubcolumnDisplay’s plot_single_profile method.
# Specify time step to examine
Time = np.array('2012-10-31T21:00', dtype='datetime64')
# generate a SubcolumnDisplay object for coords closest to the NSA site.
model_display4 = emc2.plotting.SubcolumnDisplay(my_e3sm, subplot_shape=(1, 4), figsize=(15, 5),
lat_sel=NSA_coords["lat"], lon_sel=NSA_coords["lon"],
tight_layout=True)
# Plot variables at a given time step.
for ii in range(4):
model_display4.plot_single_profile(field_to_plot[ii], time=Time, log_plot=log_plot[ii], y_range=y_range,
subplot_index=(0, ii), cmap=cmap, title=Time,
vmin=vmin_max[ii][0], vmax=vmin_max[ii][1])
cropping lat dim (lat requested = 71.32)
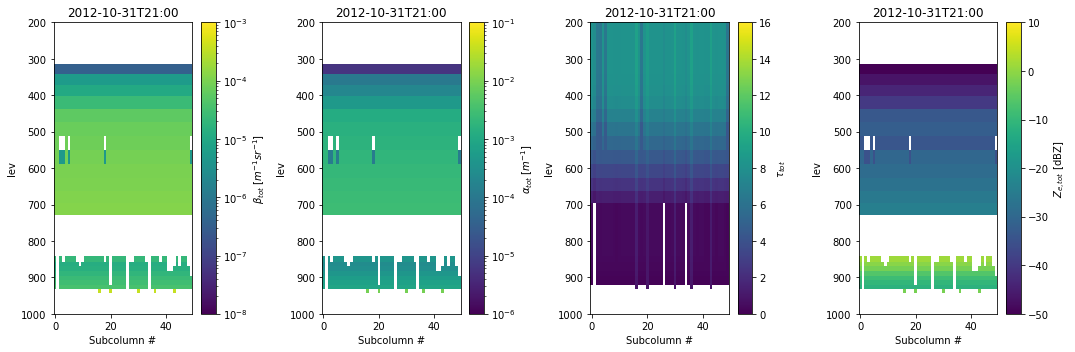
We can also examine mean profiles using the plot_subcolumn_mean_profile method. This method also illustrates the stardard deviation as an envelope around the mean. Note that here we plot the mean profile as a function of height instead of pressure coordinates by setting pressure_coords=False when invoking the plotting routine.
# Specify height range to plot (previously we plotted the pressure level on the y-axis)
y_range_h = (0., 12000)
# generate a SubcolumnDisplay object for coords closest to the NSA site
model_display5 = emc2.plotting.SubcolumnDisplay(my_e3sm, subplot_shape=(1, 4), figsize=(15, 5),
lat_sel=NSA_coords["lat"], lon_sel=NSA_coords["lon"],
tight_layout=True)
# Plot variable mean profiles
for ii in range(4):
model_display5.plot_subcolumn_mean_profile(field_to_plot[ii], time=Time, log_plot=log_plot[ii], y_range=y_range_h,
subplot_index=(0, ii), title=Time, pressure_coords=False,
x_range=vmin_max[ii])
cropping lat dim (lat requested = 71.32)

Concluding Remarks#
EMC² includes support for a large number of instruments including the full ARM radar and lidar instrument suite and has already been operated on ESM, SCM, and LES outputs. The software include multiple methods for phase classification and partitioning, scattering calculations, and more. See and follow the continuously updated EMC² GitHub repository for more information.

