Extracted Radar Columns and In-Situ Sensors (RadCLss) Dataset#
14 March 2022 Case#
In order to evaluate the SAIL field campaign snowfall retrievals, a dataset containing the radar column above each of our sites of interest, matched with in-situ ground observations is desired.
Current List of Supported In-Situ Ground Observations#
Pluvio Weighing Bucket Precipitation Gauge [WBPLUVIO2] (DOI: 10.5439/1338194)
Surface Meteorological Instrumentation [MET] (DOI: 10.5439/1786358)
Laser Disdrometer [LD] (DOI: 10.5439/1779709)
Balloon-borne sounding system [SONDEWNPN] (DOI: 10.5439/1595321)
Radar Wind Profiler [915RWPPRECIPMOMENTHIGH]
Imports#
import glob
import os
import datetime
import numpy as np
import matplotlib.pyplot as plt
import pyart
import xarray as xr
import pandas as pd
import act
from matplotlib.dates import DateFormatter
from matplotlib import colors
Define Processing Variables#
# Define the desired processing date for the CSU XPRECIPRADAR in YYYY-MM-DD format.
DATE = "2022-03-14"
# Define the directory where the CSU-X Band CMAC2.0 files are located.
RADAR_DIR = "/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/"
# Define an output directory for downloaded ground instrumentation
INSITU_DIR = '/Users/jrobrien/ARM/inProgress/'
# Define ARM Username and ARM Token with ARM Live service for downloading ground instrumentation via ACT.DISCOVERY
# With your ARM username, you can find your ARM Live token here: https://adc.arm.gov/armlive/
ARM_USERNAME = os.getenv("ARM_USERNAME")
ARM_TOKEN = os.getenv("ARM_TOKEN")
Define Functions#
def subset_points(file, lats, lons, sites):
"""Subset a radar file for a set of latitudes and longitudes"""
# Read in the file
radar = pyart.io.read(file)
column_list = []
for lat, lon in zip(lats, lons):
# Make sure we are interpolating from the radar's location above sea level
# NOTE: interpolating throughout Troposphere to match sonde to in the future
da = pyart.util.columnsect.get_field_location(radar, lat, lon).interp(height=np.arange(np.round(radar.altitude['data'][0]), 10100, 100))
# Add the latitude and longitude of the extracted column
da["latitude"], da["longitude"] = lat, lon
# Time is based off the start of the radar volume
dt = pd.to_datetime(radar.time["data"], unit='s')[-1]
da["time"] = [dt]
column_list.append(da)
# Concatenate the extracted radar columns for this scan across all sites
ds = xr.concat(column_list, dim='site')
ds["site"] = sites
# Add attributes for Time, Latitude, Longitude, and Sites
ds.time.attrs.update(long_name=('Time in Seconds that Cooresponds to the Start'
+ " of each Individual Radar Volume Scan before"
+ " Concatenation"),
description=('Time in Seconds that Cooresponds to the Minimum'
+ ' Height Gate'))
ds.site.attrs.update(long_name="SAIL/SPLASH In-Situ Ground Observation Site Identifers")
ds.latitude.attrs.update(long_name='Latitude of SAIL Ground Observation Site',
units='Degrees North')
ds.longitude.attrs.update(long_name='Longitude of SAIL Ground Observation Site',
units='Degrees East')
return ds
def match_datasets_act(column, ground, site, discard, resample='sum', DataSet=False):
"""
Time synchronization of a Ground Instrumentation Dataset to
a Radar Column for Specific Locations using the ARM ACT package
Parameters
----------
column : Xarray DataSet
Xarray DataSet containing the extracted radar column above multiple locations.
Dimensions should include Time, Height, Site
ground : str; Xarray DataSet
String containing the path of the ground instrumentation file that is desired
to be included within the extracted radar column dataset.
If DataSet is set to True, ground is Xarray Dataset and will skip I/O.
site : str
Location of the ground instrument. Should be included within the filename.
discard : list
List containing the desired input ground instrumentation variables to be
removed from the xarray DataSet.
resample : str
Mathematical operational for resampling ground instrumentation to the radar time.
Default is to sum the data across the resampling period. Checks for mean.
DataSet : boolean
Boolean flag to determine if ground input is an Xarray Dataset.
Set to True if ground input is Xarray DataSet.
Returns
-------
ds : Xarray DataSet
Xarray Dataset containing the time-synced in-situ ground observations with
the inputed radar column
"""
# Check to see if input is xarray DataSet or a file path
if DataSet == True:
grd_ds = ground
else:
# Read in the file using ACT
grd_ds = act.io.armfiles.read_netcdf(ground, cleanup_qc=True, drop_variables=discard)
# Default are Lazy Arrays; convert for matching with column
grd_ds = grd_ds.compute()
# Remove Base_Time before Resampling Data since you can't force 1 datapoint to 5 min sum
if 'base_time' in grd_ds.data_vars:
del grd_ds['base_time']
# Check to see if height is a dimension within the ground instrumentation.
# If so, first interpolate heights to match radar, before interpolating time.
if 'height' in grd_ds.dims:
grd_ds = grd_ds.interp(height=np.arange(column['height'].data[0], 10100, 100), method='linear')
# Check to see if ground instrumentation is the RWP, as it has conflicting variable names
# with the CMAC2.0 extracted radar columns
if 'signal_to_noise_ratio' in grd_ds.data_vars:
grd_ds = grd_ds.rename_vars(signal_to_noise_ratio='rwp_signal_to_noise_ratio')
# Resample the ground data to 5 min and interpolate to the CSU X-Band time.
# Keep data variable attributes to help distingish between instruments/locations
if resample.split('=')[-1] == 'mean':
matched = grd_ds.resample(time='5Min',
closed='right').mean(keep_attrs=True).interp(time=column.time,
method='linear')
else:
matched = grd_ds.resample(time='5Min',
closed='right').sum(keep_attrs=True).interp(time=column.time,
method='linear')
# Add SAIL site location as a dimension for the Pluvio data
matched = matched.assign_coords(coords=dict(site=site))
matched = matched.expand_dims('site')
# Remove Lat/Lon Data variables as it is included within the Matched Dataset with Site Identfiers
if 'lat' in matched.data_vars:
del matched['lat']
if 'lon' in matched.data_vars:
del matched['lon']
# Update the individual Variables to Hold Global Attributes
# global attributes will be lost on merging into the matched dataset.
# Need to keep as many references and descriptors as possible
for var in matched.data_vars:
matched[var].attrs.update(source=matched.datastream)
# Merge the two DataSets
column = xr.merge([column, matched])
return column
Define the Location of the SAIL Sites.#
# Define the splash locations [lon,lat]
kettle_ponds = [-106.9731488, 38.9415427]
brush_creek = [-106.920259, 38.8596282]
avery_point = [-106.9965928, 38.9705885]
pumphouse_site = [-106.9502476, 38.9226741]
roaring_judy = [-106.8530215, 38.7170576]
M1 = [-106.987, 38.9267]
snodgrass = [-106.978929, 38.926572]
sites = ["M1", "kettle_ponds", "brush_creek", "avery_point",
"pumphouse_site", "roaring_judy", "snodgrass"]
# Zip these together!
lons, lats = list(zip(M1,
kettle_ponds,
brush_creek,
avery_point,
pumphouse_site,
roaring_judy,
snodgrass))
List the Available CSU X-Band Corrected Precipitation Radar Moments in Antenna Coordinates Version 2 (CMAC2.0) Files#
In contrast to the Merged CSU X-Band files previously used, the CSU X-Band CMAC2.0 files have been corrected for beam blockage and clutter, as well as, undergone additional processing.#
Z-S Relationships previously explored are applied during the CMAC2.0 processing of the CSU X-Band data. Therefore, all we need to do now is extract the radar columns and combine with ground instruments.#
# With the user defined RADAR_DIR, grab all the XPRECIPRADAR CMAC files for the defined DATE
file_list = sorted(glob.glob(RADAR_DIR + '*' + DATE.replace('-', '') + '*.nc'))
file_list[:10]
['/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-000239.nc',
'/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-001319.nc',
'/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-003439.nc',
'/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-004519.nc',
'/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-005039.nc',
'/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-011159.nc',
'/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-013839.nc',
'/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-015439.nc',
'/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-021039.nc',
'/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-021559.nc']
%%time
ds_list = []
for file in file_list[:]:
print(file)
ds_list.append(subset_points(file, lats, lons, sites))
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-000239.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-001319.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-003439.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-004519.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-005039.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-011159.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-013839.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-015439.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-021039.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-021559.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-023159.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-023719.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-024239.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-024759.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-030920.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-032520.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-033040.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-033600.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-034120.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-034640.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-035720.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-040240.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-041320.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-041840.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-042400.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-042920.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-050120.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-053320.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-053840.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-054401.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-055440.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-061601.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-062121.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-064801.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-065321.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-070401.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-071441.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-072521.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-073041.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-073601.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-075721.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-081321.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-081841.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-082401.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-082921.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-084001.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-084521.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-090642.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-093842.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-094922.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-095442.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-101602.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-102122.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-103722.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-112002.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-112522.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-113602.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-115723.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-120243.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-120803.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-123443.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-124003.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-125043.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-125603.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-130123.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-131203.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-131723.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-132243.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-134923.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-140003.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-142123.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-153044.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-153604.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-154644.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-155724.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-161844.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-163444.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-164524.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-165044.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-170124.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-171204.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-172244.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-173324.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-173844.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-174924.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-175445.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-181044.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-182125.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-182645.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-184245.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-185325.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-185845.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-190405.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-190925.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-192005.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-192525.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-193045.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-195205.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-195725.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-200805.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-201325.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-203445.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-204005.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-205606.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-210646.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-211726.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-220006.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-220526.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-222126.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-223206.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-223726.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-224806.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-225846.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-230926.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-232006.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-232526.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-233607.nc
/Users/jrobrien/ARM/data/CSU-XPrecipRadar/cmac2/gucxprecipradarcmacM1.c1.20220314-235727.nc
CPU times: user 12min 6s, sys: 54.2 s, total: 13min
Wall time: 14min 26s
Combine all Extracted Radar Columns to Form Daily Timeseries#
# Concatenate all extracted columns across time dimension to form daily timeseries
ds = xr.concat(ds_list, dim='time')
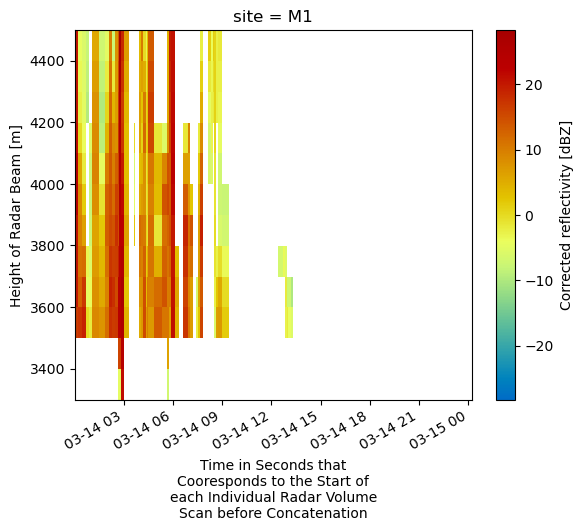
ds.sel(site='M1').sel(height=slice(3300, 4500)).corrected_reflectivity.plot(x='time',
cmap='pyart_HomeyerRainbow')
<matplotlib.collections.QuadMesh at 0x15ae18f10>
# Remove Global Attributes from the Column Extraction
# Attributes make sense for single location, but not collection of sites.
ds.attrs = {}
# Remove the Base_Time variable from extracted column
del ds['base_time']
ds
<xarray.Dataset>
Dimensions: (time: 118, site: 7, height: 70)
Coordinates:
* height (height) float64 3.149e+03 ... ...
* time (time) datetime64[ns] 2022-03-1...
* site (site) <U14 'M1' ... 'snodgrass'
Data variables: (12/34)
DBZ (time, site, height) float64 na...
VEL (time, site, height) float64 na...
WIDTH (time, site, height) float64 na...
ZDR (time, site, height) float64 na...
PHIDP (time, site, height) float64 na...
RHOHV (time, site, height) float64 na...
... ...
snow_rate_ws2012 (time, site, height) float64 na...
snow_rate_ws88diw (time, site, height) float64 na...
snow_rate_m2009_1 (time, site, height) float64 na...
snow_rate_m2009_2 (time, site, height) float64 na...
latitude (time, site) float64 38.93 ... ...
longitude (time, site) float64 -107.0 ......xarray.Dataset
- time: 118
- site: 7
- height: 70
- height(height)float643.149e+03 3.249e+03 ... 1.005e+04
- long_name :
- Height of Radar Beam
- units :
- m
- standard_name :
- height
- description :
- Height Above Sea Level [in meters] for the Center of Each Radar Gate Above the Target Location
array([ 3149., 3249., 3349., 3449., 3549., 3649., 3749., 3849., 3949., 4049., 4149., 4249., 4349., 4449., 4549., 4649., 4749., 4849., 4949., 5049., 5149., 5249., 5349., 5449., 5549., 5649., 5749., 5849., 5949., 6049., 6149., 6249., 6349., 6449., 6549., 6649., 6749., 6849., 6949., 7049., 7149., 7249., 7349., 7449., 7549., 7649., 7749., 7849., 7949., 8049., 8149., 8249., 8349., 8449., 8549., 8649., 8749., 8849., 8949., 9049., 9149., 9249., 9349., 9449., 9549., 9649., 9749., 9849., 9949., 10049.]) - time(time)datetime64[ns]2022-03-14T00:06:47 ... 2022-03-...
- long_name :
- Time in Seconds that Cooresponds to the Start of each Individual Radar Volume Scan before Concatenation
- description :
- Time in Seconds that Cooresponds to the Minimum Height Gate
array(['2022-03-14T00:06:47.000000000', '2022-03-14T00:17:28.000000000', '2022-03-14T00:38:47.000000000', '2022-03-14T00:49:28.000000000', '2022-03-14T00:54:48.000000000', '2022-03-14T01:16:08.000000000', '2022-03-14T01:42:48.000000000', '2022-03-14T01:58:48.000000000', '2022-03-14T02:14:48.000000000', '2022-03-14T02:20:08.000000000', '2022-03-14T02:36:08.000000000', '2022-03-14T02:41:28.000000000', '2022-03-14T02:46:48.000000000', '2022-03-14T02:52:08.000000000', '2022-03-14T03:13:29.000000000', '2022-03-14T03:29:28.000000000', '2022-03-14T03:34:49.000000000', '2022-03-14T03:40:09.000000000', '2022-03-14T03:45:29.000000000', '2022-03-14T03:50:49.000000000', '2022-03-14T04:01:28.000000000', '2022-03-14T04:06:49.000000000', '2022-03-14T04:17:29.000000000', '2022-03-14T04:22:49.000000000', '2022-03-14T04:28:09.000000000', '2022-03-14T04:33:29.000000000', '2022-03-14T05:05:29.000000000', '2022-03-14T05:37:29.000000000', '2022-03-14T05:42:49.000000000', '2022-03-14T05:48:09.000000000', '2022-03-14T05:58:49.000000000', '2022-03-14T06:20:10.000000000', '2022-03-14T06:25:29.000000000', '2022-03-14T06:52:10.000000000', '2022-03-14T06:57:30.000000000', '2022-03-14T07:08:10.000000000', '2022-03-14T07:18:50.000000000', '2022-03-14T07:29:30.000000000', '2022-03-14T07:34:50.000000000', '2022-03-14T07:40:10.000000000', '2022-03-14T08:01:30.000000000', '2022-03-14T08:17:30.000000000', '2022-03-14T08:22:50.000000000', '2022-03-14T08:28:10.000000000', '2022-03-14T08:33:30.000000000', '2022-03-14T08:44:10.000000000', '2022-03-14T08:49:30.000000000', '2022-03-14T09:10:51.000000000', '2022-03-14T09:42:50.000000000', '2022-03-14T09:53:30.000000000', '2022-03-14T09:58:51.000000000', '2022-03-14T10:20:11.000000000', '2022-03-14T10:25:31.000000000', '2022-03-14T10:41:31.000000000', '2022-03-14T11:24:11.000000000', '2022-03-14T11:29:31.000000000', '2022-03-14T11:40:11.000000000', '2022-03-14T12:01:31.000000000', '2022-03-14T12:06:52.000000000', '2022-03-14T12:12:11.000000000', '2022-03-14T12:38:51.000000000', '2022-03-14T12:44:11.000000000', '2022-03-14T12:54:52.000000000', '2022-03-14T13:00:12.000000000', '2022-03-14T13:05:32.000000000', '2022-03-14T13:16:12.000000000', '2022-03-14T13:21:32.000000000', '2022-03-14T13:26:52.000000000', '2022-03-14T13:53:32.000000000', '2022-03-14T14:04:13.000000000', '2022-03-14T14:25:32.000000000', '2022-03-14T15:34:52.000000000', '2022-03-14T15:40:13.000000000', '2022-03-14T15:50:53.000000000', '2022-03-14T16:01:33.000000000', '2022-03-14T16:22:53.000000000', '2022-03-14T16:38:53.000000000', '2022-03-14T16:49:33.000000000', '2022-03-14T16:54:53.000000000', '2022-03-14T17:05:33.000000000', '2022-03-14T17:16:13.000000000', '2022-03-14T17:26:53.000000000', '2022-03-14T17:37:33.000000000', '2022-03-14T17:42:53.000000000', '2022-03-14T17:53:34.000000000', '2022-03-14T17:58:53.000000000', '2022-03-14T18:14:54.000000000', '2022-03-14T18:25:34.000000000', '2022-03-14T18:30:54.000000000', '2022-03-14T18:46:54.000000000', '2022-03-14T18:57:34.000000000', '2022-03-14T19:02:53.000000000', '2022-03-14T19:08:14.000000000', '2022-03-14T19:13:34.000000000', '2022-03-14T19:24:14.000000000', '2022-03-14T19:29:34.000000000', '2022-03-14T19:34:54.000000000', '2022-03-14T19:56:14.000000000', '2022-03-14T20:01:34.000000000', '2022-03-14T20:12:14.000000000', '2022-03-14T20:17:34.000000000', '2022-03-14T20:38:54.000000000', '2022-03-14T20:44:14.000000000', '2022-03-14T21:00:15.000000000', '2022-03-14T21:10:54.000000000', '2022-03-14T21:21:34.000000000', '2022-03-14T22:04:15.000000000', '2022-03-14T22:09:35.000000000', '2022-03-14T22:25:35.000000000', '2022-03-14T22:36:15.000000000', '2022-03-14T22:41:35.000000000', '2022-03-14T22:52:15.000000000', '2022-03-14T23:02:55.000000000', '2022-03-14T23:13:35.000000000', '2022-03-14T23:24:15.000000000', '2022-03-14T23:29:35.000000000', '2022-03-14T23:40:16.000000000', '2022-03-15T00:01:36.000000000'], dtype='datetime64[ns]') - site(site)<U14'M1' 'kettle_ponds' ... 'snodgrass'
- long_name :
- SAIL/SPLASH In-Situ Ground Observation Site Identifers
array(['M1', 'kettle_ponds', 'brush_creek', 'avery_point', 'pumphouse_site', 'roaring_judy', 'snodgrass'], dtype='<U14')
- DBZ(time, site, height)float64nan -10.82 -6.316 ... nan nan nan
- units :
- dBZ
- standard_name :
- equivalent_reflectivity_factor
- coordinates :
- elevation azimuth range
array([[[ nan, -10.81999969, -6.31648953, ..., nan, nan, nan], [ nan, 16.10779893, 3.05952826, ..., nan, nan, nan], [ nan, -12.97489854, -11.73341511, ..., nan, nan, nan], ..., [ nan, 7.30825315, 4.06749179, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, -10.93107905, -4.84339817, ..., nan, nan, nan]], [[ nan, -10.81999969, -5.41896141, ..., nan, nan, nan], [ nan, 13.24047607, 19.11138189, ..., nan, nan, nan], [ nan, -12.23780284, -15.16765685, ..., nan, nan, nan], ... [ nan, -15.92000008, -15.92000008, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, -11.84000015, -11.80514278, ..., nan, nan, nan]], [[ nan, -10.81999969, -10.81999969, ..., nan, nan, nan], [ nan, -9.90999985, -10.14783067, ..., nan, nan, nan], [ nan, -20.18759306, -11.14999962, ..., nan, nan, nan], ..., [ nan, -15.92000008, -15.92000008, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 7.10398186, -11.84000015, ..., nan, nan, nan]]]) - VEL(time, site, height)float64nan 12.41 -10.28 ... nan nan nan
- units :
- m/s
- standard_name :
- radial_velocity_of_scatterers_away_from_instruments
- coordinates :
- elevation azimuth range
array([[[ nan, 12.40704339, -10.28064785, ..., nan, nan, nan], [ nan, 0.53648465, 20.08120593, ..., nan, nan, nan], [ nan, -22.87388508, -2.03048569, ..., nan, nan, nan], ..., [ nan, -2.02765943, -1.73831865, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, -22.50603496, -3.05127251, ..., nan, nan, nan]], [[ nan, 12.16807962, -9.74842282, ..., nan, nan, nan], [ nan, -0.62554882, -1.47551063, ..., nan, nan, nan], [ nan, -21.83998237, 3.957761 , ..., nan, nan, nan], ... [ nan, -30.4364079 , 0.97140561, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, -18.75076925, -18.59143882, ..., nan, nan, nan]], [[ nan, 11.12294777, -6.95099764, ..., nan, nan, nan], [ nan, -11.09767179, -0.32891421, ..., nan, nan, nan], [ nan, 4.73620301, 3.23004115, ..., nan, nan, nan], ..., [ nan, 21.93758999, -19.01108051, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, -26.08425652, -3.89201876, ..., nan, nan, nan]]]) - WIDTH(time, site, height)float64nan -9.99e+04 ... nan nan
- units :
- m/s
- standard_name :
- doppler_spectrum_width
- coordinates :
- elevation azimuth range
array([[[ nan, -9.99000000e+04, -8.22842531e+04, ..., nan, nan, nan], [ nan, -4.08106716e+03, -9.99000000e+04, ..., nan, nan, nan], [ nan, 2.50910034e+00, 3.06551213e+00, ..., nan, nan, nan], ..., [ nan, 7.58765939e-01, 7.29957409e-01, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, -9.99000000e+04, -9.99000000e+04, ..., nan, nan, nan]], [[ nan, -9.99000000e+04, -8.22843782e+04, ..., nan, nan, nan], [ nan, 1.01529010e+00, 6.68276547e-01, ..., nan, nan, nan], [ nan, 2.60999932e+00, 2.02195144e+00, ..., nan, nan, nan], ... [ nan, 3.29708401e+00, 1.78712845e+00, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, -3.07787582e+04, -9.99000000e+04, ..., nan, nan, nan]], [[ nan, -9.99000000e+04, -9.99000000e+04, ..., nan, nan, nan], [ nan, -9.58179381e+04, 1.88353219e+00, ..., nan, nan, nan], [ nan, 2.38710085e+00, 3.38478018e+00, ..., nan, nan, nan], ..., [ nan, 2.84406397e+00, 3.18489355e+00, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, -9.99000000e+04, -9.99000000e+04, ..., nan, nan, nan]]]) - ZDR(time, site, height)float64nan 1.723 1.945 ... nan nan nan
- units :
- dB
- standard_name :
- log_differential_reflectivity_hv
- coordinates :
- elevation azimuth range
array([[[ nan, 1.72313261e+00, 1.94521916e+00, ..., nan, nan, nan], [ nan, 2.64077369e+00, 1.66025513e+00, ..., nan, nan, nan], [ nan, 2.52790001e+00, 2.35619493e+00, ..., nan, nan, nan], ..., [ nan, 2.30185108e+00, 2.34631925e+00, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 7.39442602e+00, 1.33644228e+00, ..., nan, nan, nan]], [[ nan, -1.54023688e+04, -8.22815469e+04, ..., nan, nan, nan], [ nan, 2.56346229e+00, 2.40023401e+00, ..., nan, nan, nan], [ nan, 2.31930082e+00, 3.17253632e+00, ..., nan, nan, nan], ... [ nan, 3.08631870e+00, 2.26874471e+00, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 3.41713546e+00, 4.01083139e+00, ..., nan, nan, nan]], [[ nan, -1.54012609e+04, -9.98969500e+04, ..., nan, nan, nan], [ nan, 3.56772003e+00, 2.83642566e+00, ..., nan, nan, nan], [ nan, 2.81229978e+00, 2.71970760e+00, ..., nan, nan, nan], ..., [ nan, 2.72776620e+00, 3.14497870e+00, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, -6.91175989e+04, 1.64093429e+00, ..., nan, nan, nan]]]) - PHIDP(time, site, height)float64nan 128.3 66.94 ... nan nan nan
- units :
- degrees
- standard_name :
- differential_phase_hv
- coordinates :
- elevation azimuth range
array([[[ nan, 128.32599258, 66.94012996, ..., nan, nan, nan], [ nan, 104.15218989, 98.95949075, ..., nan, nan, nan], [ nan, 327.50664688, 274.91695297, ..., nan, nan, nan], ..., [ nan, 100.52865903, 100.0267886 , ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 233.82960237, 101.40374094, ..., nan, nan, nan]], [[ nan, 102.38336946, 124.70658576, ..., nan, nan, nan], [ nan, 100.82576292, 101.78166027, ..., nan, nan, nan], [ nan, 98.14142405, 210.7710162 , ..., nan, nan, nan], ... [ nan, 353.77016448, 342.00584653, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 61.95795905, 13.99077346, ..., nan, nan, nan]], [[ nan, 79.69283065, 140.7436992 , ..., nan, nan, nan], [ nan, 246.09510138, 139.59054037, ..., nan, nan, nan], [ nan, 315.84865017, 212.66277956, ..., nan, nan, nan], ..., [ nan, 292.30180737, 231.35807351, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 85.45987434, 203.35780229, ..., nan, nan, nan]]]) - RHOHV(time, site, height)float64nan 0.8448 0.8739 ... nan nan nan
- units :
- 1
- standard_name :
- cross_correlation_ratio_hv
- coordinates :
- elevation azimuth range
array([[[ nan, 8.44771128e-01, 8.73862805e-01, ..., nan, nan, nan], [ nan, 9.94877481e-01, 9.89414924e-01, ..., nan, nan, nan], [ nan, 2.99380072e-01, 4.13956193e-01, ..., nan, nan, nan], ..., [ nan, 9.95876644e-01, 9.94376618e-01, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 6.14005694e-01, 9.13857231e-01, ..., nan, nan, nan]], [[ nan, 9.40221613e-01, 7.88435599e-01, ..., nan, nan, nan], [ nan, 9.96081772e-01, 9.98057476e-01, ..., nan, nan, nan], [ nan, 5.34169890e-01, 3.32643878e-01, ..., nan, nan, nan], ... [ nan, 3.95857605e-01, 5.99340293e-01, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 6.16037804e-01, 5.30085697e-01, ..., nan, nan, nan]], [[ nan, 9.84233782e-01, 9.66666287e-01, ..., nan, nan, nan], [ nan, 7.24832237e-01, 6.07525477e-01, ..., nan, nan, nan], [ nan, 6.52559980e-01, 5.35356055e-01, ..., nan, nan, nan], ..., [ nan, 5.86753201e-01, 6.05972443e-01, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 7.96713342e-01, 5.25898647e-01, ..., nan, nan, nan]]]) - NCP(time, site, height)float64nan 0.97 0.9612 ... nan nan nan
- units :
- 1
- standard_name :
- normalized_coherent_power
- coordinates :
- elevation azimuth range
array([[[ nan, 0.96999997, 0.96118338, ..., nan, nan, nan], [ nan, 0.94081721, 0.96057445, ..., nan, nan, nan], [ nan, 0.11420001, 0.14346343, ..., nan, nan, nan], ..., [ nan, 0.95999998, 0.9612553 , ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 0.96999997, 0.96683114, ..., nan, nan, nan]], [[ nan, 0.96999997, 0.96999997, ..., nan, nan, nan], [ nan, 0.9508172 , 0.96999997, ..., nan, nan, nan], [ nan, 0.10789999, 0.09999999, ..., nan, nan, nan], ... [ nan, 0.11123405, 0.13251064, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 0.96999997, 0.96683114, ..., nan, nan, nan]], [[ nan, 0.96999997, 0.93826027, ..., nan, nan, nan], [ nan, 0.64774175, 0.35023398, ..., nan, nan, nan], [ nan, 0.11949997, 0.08897562, ..., nan, nan, nan], ..., [ nan, 0.11 , 0.1049787 , ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 0.96999997, 0.96674664, ..., nan, nan, nan]]]) - DBZhv(time, site, height)float64nan 5.768 15.82 ... nan nan nan
- units :
- dBZ
- standard_name :
- equivalent_reflectivity_factor_hv
- coordinates :
- elevation azimuth range
array([[[ nan, 5.76836845e+00, 1.58196087e+01, ..., nan, nan, nan], [ nan, 1.49226638e+01, 9.95344338e+00, ..., nan, nan, nan], [ nan, -1.82728000e+01, -1.78696588e+01, ..., nan, nan, nan], ..., [ nan, 7.66961466e+00, 4.31130019e+00, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 4.88642299e+00, -1.81306186e+00, ..., nan, nan, nan]], [[ nan, 9.12104147e+00, 1.74641430e+01, ..., nan, nan, nan], [ nan, 1.39973789e+01, 1.91350430e+01, ..., nan, nan, nan], [ nan, -2.18722988e+01, -2.06688283e+01, ..., nan, nan, nan], ... [ nan, -2.37485994e+01, -2.94660406e+01, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 5.72187284e+00, -1.59477157e+01, ..., nan, nan, nan]], [[ nan, 9.74759839e+00, 1.57818002e+01, ..., nan, nan, nan], [ nan, -1.34555053e+01, -1.36941074e+01, ..., nan, nan, nan], [ nan, -1.73216009e+01, -1.97473659e+01, ..., nan, nan, nan], ..., [ nan, -2.85639758e+01, -2.36445975e+01, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 1.58022412e+01, -1.66800558e+01, ..., nan, nan, nan]]]) - cbb_flag(time, site, height)float64nan 0.8458 0.0 0.0 ... nan nan nan
- units :
- 1
- long_name :
- Cumulative Beam Block Fraction Flag
- coordinates :
- elevation azimuth range
array([[[ nan, 0.84578254, 0. , ..., nan, nan, nan], [ nan, 0. , 0. , ..., nan, nan, nan], [ nan, 1. , 1. , ..., nan, nan, nan], ..., [ nan, 0. , 0. , ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 0. , 0. , ..., nan, nan, nan]], [[ nan, 0.84578254, 0. , ..., nan, nan, nan], [ nan, 0. , 0. , ..., nan, nan, nan], [ nan, 1. , 1. , ..., nan, nan, nan], ... [ nan, 0. , 0. , ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 0. , 0. , ..., nan, nan, nan]], [[ nan, 0.84578254, 0. , ..., nan, nan, nan], [ nan, 0. , 0. , ..., nan, nan, nan], [ nan, 1. , 1. , ..., nan, nan, nan], ..., [ nan, 0. , 0. , ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 0. , 0. , ..., nan, nan, nan]]]) - sounding_temperature(time, site, height)float64nan -3.025 -4.007 ... nan nan nan
- units :
- degC
- standard_name :
- interpolated_profile
- long_name :
- Interpolated profile
array([[[ nan, -3.02489149, -4.00664871, ..., nan, nan, nan], [ nan, -3.00644346, -4.00498955, ..., nan, nan, nan], [ nan, -3.0312439 , -3.99862265, ..., nan, nan, nan], ..., [ nan, -3.0108487 , -4.00585198, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, -3.03686548, -4.00033004, ..., nan, nan, nan]], [[ nan, -3.02489149, -4.00664871, ..., nan, nan, nan], [ nan, -3.00644346, -4.00498955, ..., nan, nan, nan], [ nan, -3.0312439 , -3.99862265, ..., nan, nan, nan], ... [ nan, -3.2624767 , -4.20459192, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, -3.24478044, -4.21020775, ..., nan, nan, nan]], [[ nan, -3.25761667, -4.22537902, ..., nan, nan, nan], [ nan, -3.26517108, -4.20358836, ..., nan, nan, nan], [ nan, -3.24960372, -4.22534105, ..., nan, nan, nan], ..., [ nan, -3.2624767 , -4.20459192, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, -3.24478044, -4.21064839, ..., nan, nan, nan]]]) - signal_to_noise_ratio(time, site, height)float64nan 0.0 4.504 19.62 ... nan nan nan
- units :
- dB
- standard_name :
- signal_to_noise_ratio
- coordinates :
- elevation azimuth range
array([[[ nan, 0. , 4.50351016, ..., nan, nan, nan], [ nan, 26.02739037, 12.96952766, ..., nan, nan, nan], [ nan, -1.82489854, -0.58341539, ..., nan, nan, nan], ..., [ nan, 23.23763593, 19.9887468 , ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 0.90892088, 6.9617442 , ..., nan, nan, nan]], [[ nan, 0. , 5.40280164, ..., nan, nan, nan], [ nan, 23.15047592, 29.02195622, ..., nan, nan, nan], [ nan, -1.08570317, -4.00990096, ..., nan, nan, nan], ... [ nan, 0. , 0. , ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 0. , 0. , ..., nan, nan, nan]], [[ nan, 0. , 0. , ..., nan, nan, nan], [ nan, 0. , -0.2372564 , ..., nan, nan, nan], [ nan, -9.02969251, 0. , ..., nan, nan, nan], ..., [ nan, 0. , 0. , ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 18.95090042, 0. , ..., nan, nan, nan]]]) - velocity_texture(time, site, height)float64nan 4.348 2.363 ... nan nan nan
- units :
- m/s
- standard_name :
- radial_velocity_of_scatterers_away_from_instrument
- long_name :
- Mean dopper velocity
- coordinates :
- elevation azimuth range
array([[[ nan, 4.34760833, 2.36289549, ..., nan, nan, nan], [ nan, 2.301518 , 4.61680673, ..., nan, nan, nan], [ nan, 5.22938283, 4.97800901, ..., nan, nan, nan], ..., [ nan, 0.6256861 , 0.64153505, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 4.18866136, 3.44449815, ..., nan, nan, nan]], [[ nan, 4.24774887, 2.32391028, ..., nan, nan, nan], [ nan, 0.33656745, 0.28420224, ..., nan, nan, nan], [ nan, 5.1155082 , 6.25700009, ..., nan, nan, nan], ... [ nan, 5.55066581, 4.61873211, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 5.12399423, 6.97992035, ..., nan, nan, nan]], [[ nan, 4.33351278, 5.17077991, ..., nan, nan, nan], [ nan, 3.48014308, 2.76671444, ..., nan, nan, nan], [ nan, 5.3655372 , 5.48115689, ..., nan, nan, nan], ..., [ nan, 5.62090783, 6.32464634, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 4.52300875, 5.60383135, ..., nan, nan, nan]]]) - gate_id(time, site, height)float64nan 5.537 2.824 ... nan nan nan
- units :
- long_name :
- Classification of dominant scatterer
- valid_max :
- 6
- valid_min :
- 0.0
array([[[ nan, 5.53734763, 2.82366835, ..., nan, nan, nan], [ nan, 0. , 0.11489414, ..., nan, nan, nan], [ nan, 6. , 6. , ..., nan, nan, nan], ..., [ nan, 1.87659471, 0.25106487, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 3. , 2.68311657, ..., nan, nan, nan]], [[ nan, 5.53734763, 2.82366835, ..., nan, nan, nan], [ nan, 2. , 2. , ..., nan, nan, nan], [ nan, 6. , 6. , ..., nan, nan, nan], ... [ nan, 3. , 3. , ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 3. , 3. , ..., nan, nan, nan]], [[ nan, 5.53734763, 3. , ..., nan, nan, nan], [ nan, 3. , 3. , ..., nan, nan, nan], [ nan, 6. , 6. , ..., nan, nan, nan], ..., [ nan, 3. , 3. , ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 3. , 3. , ..., nan, nan, nan]]]) - simulated_velocity(time, site, height)float64nan -3.519 -2.875 ... nan nan nan
- units :
- m/s
- standard_name :
- radial_velocity_of_scatterers_away_from_instrument
- long_name :
- Simulated mean doppler velocity
- coordinates :
- elevation azimuth range
array([[[ nan, -3.51907816, -2.87505424, ..., nan, nan, nan], [ nan, -3.96451906, -3.28795881, ..., nan, nan, nan], [ nan, 3.93434608, 3.14877457, ..., nan, nan, nan], ..., [ nan, -3.90425293, -3.32425956, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, -3.68023146, -2.94900991, ..., nan, nan, nan]], [[ nan, -3.52440266, -2.87806457, ..., nan, nan, nan], [ nan, -3.96362425, -3.28847755, ..., nan, nan, nan], [ nan, 3.93466513, 3.1488073 , ..., nan, nan, nan], ... [ nan, -5.39031814, -5.59989327, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, -5.93915884, -5.81107424, ..., nan, nan, nan]], [[ nan, -5.74909365, -5.65848637, ..., nan, nan, nan], [ nan, -5.83824951, -5.92616895, ..., nan, nan, nan], [ nan, 5.79236812, 5.85058195, ..., nan, nan, nan], ..., [ nan, -5.39203247, -5.59354396, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, -5.94086844, -5.81614524, ..., nan, nan, nan]]]) - corrected_velocity(time, site, height)float64nan nan nan nan ... nan nan nan nan
- units :
- m/s
- standard_name :
- corrected_radial_velocity_of_scatterers_away_from_instrument
- long_name :
- Corrected mean doppler velocity
- valid_max :
- 47.7
- valid_min :
- -47.7
- coordinates :
- elevation azimuth range
array([[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, -0.62554882, -1.47551063, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - unfolded_differential_phase(time, site, height)float64nan 0.0 0.0 0.0 ... nan nan nan nan
- units :
- degrees
- standard_name :
- differential_phase_hv
- long_name :
- Unfolded differential propagation phase shift
- coordinates :
- elevation azimuth range
array([[[nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], ..., [nan, 0., 0., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan]], [[nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], ..., [nan, 0., 0., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan]], [[nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], ..., ... ..., [nan, 0., 0., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan]], [[nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], ..., [nan, 0., 0., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan]], [[nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], ..., [nan, 0., 0., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan]]]) - corrected_differential_phase(time, site, height)float64nan 0.01 0.01 0.01 ... nan nan nan
- units :
- degrees
- standard_name :
- differential_phase_hv
- long_name :
- Corrected differential propagation phase shift
- valid_max :
- 400.0
- valid_min :
- 0.0
- coordinates :
- elevation azimuth range
array([[[ nan, 0.01, 0.01, ..., nan, nan, nan], [ nan, 0.01, 0.01, ..., nan, nan, nan], [ nan, 0.01, 0.01, ..., nan, nan, nan], ..., [ nan, 0.01, 0.01, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 0.01, 0.01, ..., nan, nan, nan]], [[ nan, 0.01, 0.01, ..., nan, nan, nan], [ nan, 0.01, 0.01, ..., nan, nan, nan], [ nan, 0.01, 0.01, ..., nan, nan, nan], ..., [ nan, 0.01, 0.01, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 0.01, 0.01, ..., nan, nan, nan]], [[ nan, 0.01, 0.01, ..., nan, nan, nan], [ nan, 0.01, 0.01, ..., nan, nan, nan], [ nan, 0.01, 0.01, ..., nan, nan, nan], ..., ... ..., [ nan, 0.01, 0.01, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 0.01, 0.01, ..., nan, nan, nan]], [[ nan, 0.01, 0.01, ..., nan, nan, nan], [ nan, 0.01, 0.01, ..., nan, nan, nan], [ nan, 0.01, 0.01, ..., nan, nan, nan], ..., [ nan, 0.01, 0.01, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 0.01, 0.01, ..., nan, nan, nan]], [[ nan, 0.01, 0.01, ..., nan, nan, nan], [ nan, 0.01, 0.01, ..., nan, nan, nan], [ nan, 0.01, 0.01, ..., nan, nan, nan], ..., [ nan, 0.01, 0.01, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, 0.01, 0.01, ..., nan, nan, nan]]]) - filtered_corrected_differential_phase(time, site, height)float64nan nan nan nan ... nan nan nan nan
- units :
- degrees
- standard_name :
- differential_phase_hv
- long_name :
- Filtered Corrected Differential Phase
- valid_max :
- 400.0
- valid_min :
- 0.0
- coordinates :
- elevation azimuth range
array([[[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], ..., ... ..., [nan, 0., 0., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan]], [[nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], ..., [nan, 0., 0., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan]], [[nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], ..., [nan, 0., 0., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan]]]) - corrected_specific_diff_phase(time, site, height)float64nan 0.0 0.0 0.0 ... nan nan nan nan
- units :
- degrees/km
- standard_name :
- specific_differential_phase_hv
- long_name :
- Specific differential phase (KDP)
- coordinates :
- elevation azimuth range
array([[[nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], ..., [nan, 0., 0., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan]], [[nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], ..., [nan, 0., 0., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan]], [[nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], ..., ... ..., [nan, 0., 0., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan]], [[nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], ..., [nan, 0., 0., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan]], [[nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], ..., [nan, 0., 0., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan]]]) - filtered_corrected_specific_diff_phase(time, site, height)float64nan 0.0 0.0 0.0 ... nan nan nan nan
- units :
- degrees/km
- standard_name :
- specific_differential_phase_hv
- long_name :
- Filtered Corrected Specific differential phase (KDP)
- coordinates :
- elevation azimuth range
array([[[nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], ..., [nan, 0., 0., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan]], [[nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], ..., [nan, 0., 0., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan]], [[nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], ..., ... ..., [nan, 0., 0., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan]], [[nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], ..., [nan, 0., 0., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan]], [[nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], ..., [nan, 0., 0., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan]]]) - corrected_differential_reflectivity(time, site, height)float64nan nan nan nan ... nan nan nan nan
- units :
- dB
- standard_name :
- corrected_log_differential_reflectivity_hv
- long_name :
- Corrected differential reflectivity
- coordinates :
- elevation azimuth range
array([[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, 2.56346229, 2.40023401, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - corrected_reflectivity(time, site, height)float64nan nan nan nan ... nan nan nan nan
- units :
- dBZ
- standard_name :
- corrected_equivalent_reflectivity_factor
- long_name :
- Corrected reflectivity
- coordinates :
- elevation azimuth range
array([[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, 13.24047607, 19.11138189, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - height_over_iso0(time, site, height)float64nan nan nan nan ... nan nan nan nan
- units :
- m
- standard_name :
- height
- long_name :
- Height of radar beam over freezing level
array([[[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., ... ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]]) - specific_attenuation(time, site, height)float64nan nan nan nan ... nan nan nan nan
- units :
- dB/km
- standard_name :
- specific_attenuation
- long_name :
- Specific attenuation
- valid_max :
- 1.0
- valid_min :
- 0.0
- coordinates :
- elevation azimuth range
array([[[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, 0., ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., ... ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]]) - path_integrated_attenuation(time, site, height)float64nan nan nan nan ... nan nan nan nan
- units :
- dB
- long_name :
- Path Integrated Attenuation
- coordinates :
- elevation azimuth range
array([[[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, 0., ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., ... ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]]) - specific_differential_attenuation(time, site, height)float64nan nan nan nan ... nan nan nan nan
- units :
- dB/km
- long_name :
- Specific Differential Attenuation
- coordinates :
- elevation azimuth range
array([[[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, 0., ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., ... ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]]) - path_integrated_differential_attenuation(time, site, height)float64nan nan nan nan ... nan nan nan nan
- units :
- dB
- long_name :
- Path Integrated Differential Attenuation
- coordinates :
- elevation azimuth range
array([[[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, 0., ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., ... ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]]) - rain_rate_A(time, site, height)float64nan nan nan nan ... nan nan nan nan
- units :
- mm/hr
- standard_name :
- rainfall_rate
- long_name :
- rainfall_rate
- valid_max :
- 400.0
- valid_min :
- 0.0
- coordinates :
- elevation azimuth range
array([[[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, 0., ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, 0., 0., ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., ... ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]]) - snow_rate_ws2012(time, site, height)float64nan nan nan nan ... nan nan nan nan
- units :
- mm/h
- standard_name :
- snowfall_rate
- long_name :
- Snowfall rate from Z using Wolf and Snider (2012)
- valid_max :
- 500
- valid_min :
- 0
- coordinates :
- elevation azimuth range
array([[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, 6.07163114, 11.79107443, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - snow_rate_ws88diw(time, site, height)float64nan nan nan nan ... nan nan nan nan
- units :
- mm/h
- standard_name :
- snowfall_rate
- long_name :
- Snowfall rate from Z using WSR 88D High Plains
- valid_max :
- 500
- valid_min :
- 0
- coordinates :
- elevation azimuth range
array([[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, 10.06866118, 19.55328488, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - snow_rate_m2009_1(time, site, height)float64nan nan nan nan ... nan nan nan nan
- units :
- mm/h
- standard_name :
- snowfall_rate
- long_name :
- Snowfall rate from Z using Matrosov et al.(2009) Braham(1990) 1
- valid_max :
- 500
- valid_min :
- 0
- coordinates :
- elevation azimuth range
array([[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, 5.74404754, 15.96398841, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - snow_rate_m2009_2(time, site, height)float64nan nan nan nan ... nan nan nan nan
- units :
- mm/h
- standard_name :
- snowfall_rate
- long_name :
- Snowfall rate from Z using Matrosov et al.(2009) Braham(1990) 2
- valid_max :
- 500
- valid_min :
- 0
- coordinates :
- elevation azimuth range
array([[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, 4.18400756, 10.76009843, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - latitude(time, site)float6438.93 38.94 38.86 ... 38.72 38.93
- long_name :
- Latitude of SAIL Ground Observation Site
- units :
- Degrees North
array([[38.9267 , 38.9415427, 38.8596282, 38.9705885, 38.9226741, 38.7170576, 38.926572 ], [38.9267 , 38.9415427, 38.8596282, 38.9705885, 38.9226741, 38.7170576, 38.926572 ], [38.9267 , 38.9415427, 38.8596282, 38.9705885, 38.9226741, 38.7170576, 38.926572 ], [38.9267 , 38.9415427, 38.8596282, 38.9705885, 38.9226741, 38.7170576, 38.926572 ], [38.9267 , 38.9415427, 38.8596282, 38.9705885, 38.9226741, 38.7170576, 38.926572 ], [38.9267 , 38.9415427, 38.8596282, 38.9705885, 38.9226741, 38.7170576, 38.926572 ], [38.9267 , 38.9415427, 38.8596282, 38.9705885, 38.9226741, 38.7170576, 38.926572 ], [38.9267 , 38.9415427, 38.8596282, 38.9705885, 38.9226741, 38.7170576, 38.926572 ], [38.9267 , 38.9415427, 38.8596282, 38.9705885, 38.9226741, 38.7170576, 38.926572 ], [38.9267 , 38.9415427, 38.8596282, 38.9705885, 38.9226741, 38.7170576, 38.926572 ], ... [38.9267 , 38.9415427, 38.8596282, 38.9705885, 38.9226741, 38.7170576, 38.926572 ], [38.9267 , 38.9415427, 38.8596282, 38.9705885, 38.9226741, 38.7170576, 38.926572 ], [38.9267 , 38.9415427, 38.8596282, 38.9705885, 38.9226741, 38.7170576, 38.926572 ], [38.9267 , 38.9415427, 38.8596282, 38.9705885, 38.9226741, 38.7170576, 38.926572 ], [38.9267 , 38.9415427, 38.8596282, 38.9705885, 38.9226741, 38.7170576, 38.926572 ], [38.9267 , 38.9415427, 38.8596282, 38.9705885, 38.9226741, 38.7170576, 38.926572 ], [38.9267 , 38.9415427, 38.8596282, 38.9705885, 38.9226741, 38.7170576, 38.926572 ], [38.9267 , 38.9415427, 38.8596282, 38.9705885, 38.9226741, 38.7170576, 38.926572 ], [38.9267 , 38.9415427, 38.8596282, 38.9705885, 38.9226741, 38.7170576, 38.926572 ], [38.9267 , 38.9415427, 38.8596282, 38.9705885, 38.9226741, 38.7170576, 38.926572 ]]) - longitude(time, site)float64-107.0 -107.0 ... -106.9 -107.0
- long_name :
- Longitude of SAIL Ground Observation Site
- units :
- Degrees East
array([[-106.987 , -106.9731488, -106.920259 , -106.9965928, -106.9502476, -106.8530215, -106.978929 ], [-106.987 , -106.9731488, -106.920259 , -106.9965928, -106.9502476, -106.8530215, -106.978929 ], [-106.987 , -106.9731488, -106.920259 , -106.9965928, -106.9502476, -106.8530215, -106.978929 ], [-106.987 , -106.9731488, -106.920259 , -106.9965928, -106.9502476, -106.8530215, -106.978929 ], [-106.987 , -106.9731488, -106.920259 , -106.9965928, -106.9502476, -106.8530215, -106.978929 ], [-106.987 , -106.9731488, -106.920259 , -106.9965928, -106.9502476, -106.8530215, -106.978929 ], [-106.987 , -106.9731488, -106.920259 , -106.9965928, -106.9502476, -106.8530215, -106.978929 ], [-106.987 , -106.9731488, -106.920259 , -106.9965928, -106.9502476, -106.8530215, -106.978929 ], [-106.987 , -106.9731488, -106.920259 , -106.9965928, -106.9502476, -106.8530215, -106.978929 ], [-106.987 , -106.9731488, -106.920259 , -106.9965928, -106.9502476, -106.8530215, -106.978929 ], ... [-106.987 , -106.9731488, -106.920259 , -106.9965928, -106.9502476, -106.8530215, -106.978929 ], [-106.987 , -106.9731488, -106.920259 , -106.9965928, -106.9502476, -106.8530215, -106.978929 ], [-106.987 , -106.9731488, -106.920259 , -106.9965928, -106.9502476, -106.8530215, -106.978929 ], [-106.987 , -106.9731488, -106.920259 , -106.9965928, -106.9502476, -106.8530215, -106.978929 ], [-106.987 , -106.9731488, -106.920259 , -106.9965928, -106.9502476, -106.8530215, -106.978929 ], [-106.987 , -106.9731488, -106.920259 , -106.9965928, -106.9502476, -106.8530215, -106.978929 ], [-106.987 , -106.9731488, -106.920259 , -106.9965928, -106.9502476, -106.8530215, -106.978929 ], [-106.987 , -106.9731488, -106.920259 , -106.9965928, -106.9502476, -106.8530215, -106.978929 ], [-106.987 , -106.9731488, -106.920259 , -106.9965928, -106.9502476, -106.8530215, -106.978929 ], [-106.987 , -106.9731488, -106.920259 , -106.9965928, -106.9502476, -106.8530215, -106.978929 ]])
Define Dictionary of Variables to Remove from Each Datastream#
discard_var = {'LD' : ['base_time', 'time_offset', 'equivalent_radar_reflectivity_ott',
'laserband_amplitude', 'qc_equivalent_radar_reflectivity_ott',
'qc_laserband_amplitude', 'sensor_temperature',
'heating_current', 'qc_heating_current', 'sensor_voltage',
'qc_sensor_voltage', 'moment1', 'moment2', 'moment3', 'moment4',
'moment5', 'moment6', 'lat', 'lon', 'alt'
],
'Pluvio' : ['base_time', 'time_offset', 'load_cell_temp', 'heater_status',
'elec_unit_temp', 'supply_volts', 'orifice_temp', 'volt_min',
'ptemp', 'lat', 'lon', 'alt'
],
'Met' : ['base_time', 'time_offset', 'time_bounds', 'logger_volt', 'qc_logger_volt',
'logger_temp', 'qc_logger_temp', 'lat', 'lon', 'alt'
],
'Sonde' : ['base_time', 'time_offset', 'lat', 'lon'],
'RWP' : ['base_time', 'time_offset', 'time_bounds', 'height_bounds',
'lat', 'lon', 'alt'
]
}
1) Add in the Pluvio Weighing Bucket Data#
# We can use the ACT module for downloading data from the ARM web service
if ARM_USERNAME is None or ARM_TOKEN is None or len(ARM_USERNAME) == 0 or len(ARM_TOKEN) == 0:
print("ACT Username and Token Not Found ")
else:
results = act.discovery.download_data(ARM_USERNAME,
ARM_TOKEN,
'gucwbpluvio2M1.a1',
DATE,
DATE,
output=INSITU_DIR)
[DOWNLOADING] gucwbpluvio2M1.a1.20220314.000000.nc
# Define the file path
npluvio = sorted(glob.glob(INSITU_DIR + '*gucwbpluvio2M1.a1.' +DATE.replace('-','') + '*'))
# Define the site location based on the filename
pluv_site = npluvio[0].split('gucwbpluvio2')[-1].split('.')[0]
# Call Match Datasets ACT
ds = match_datasets_act(ds, npluvio[0], pluv_site, discard=discard_var['Pluvio'])
# Plot the Weighing Bucket accumulation at the M1 site
# Test to make sure Pluvio is correctly merged in with extracted column.
ds.sel(site='M1').bucket_rt.plot(x='time')
[<matplotlib.lines.Line2D at 0x159ddc5b0>]

2) Add the Surface Meteorological Station (MET) to the Matched Dataset#
# We can use the ACT module for downloading data from the ARM web service
if ARM_USERNAME is None or ARM_TOKEN is None or len(ARM_USERNAME) == 0 or len(ARM_TOKEN) == 0:
print("ACT Username and Token Not Found ")
else:
results = act.discovery.download_data(ARM_USERNAME,
ARM_TOKEN,
'gucmetM1.b1',
DATE,
DATE,
output=INSITU_DIR)
[DOWNLOADING] gucmetM1.b1.20220314.000000.cdf
# Define the file path
nmet = sorted(glob.glob(INSITU_DIR + '*gucmetM1.b1.' + DATE.replace('-', '') + '*'))
# Define the site location based on the filename
met_site = nmet[0].split('gucmet')[-1].split('.')[0]
# Call Match Datasets ACT
ds = match_datasets_act(ds, nmet[0], met_site, discard=discard_var['Met'])
# Plot the MET accumulation at the M1 site
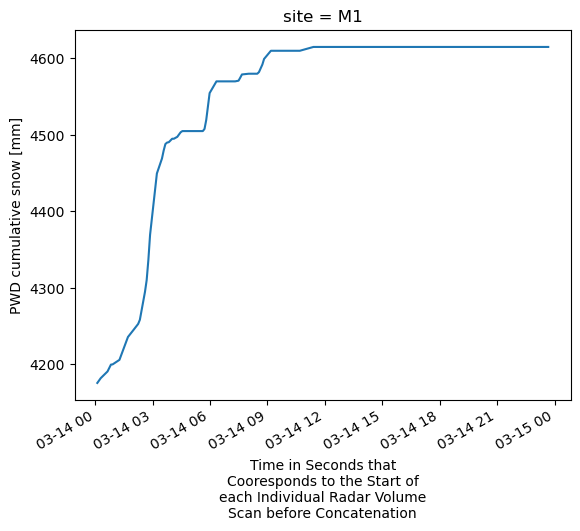
# Test to make sure MET is correctly merged in with extracted column.
ds.sel(site='M1').pwd_cumul_snow.plot(x='time')
[<matplotlib.lines.Line2D at 0x159de7d30>]
3) Add the Laser Disdrometer to the Matched Dataset - M1 Sites#
# We can use the ACT module for downloading data from the ARM web service
if ARM_USERNAME is None or ARM_TOKEN is None or len(ARM_USERNAME) == 0 or len(ARM_TOKEN) == 0:
print("ACT Username and Token Not Found ")
else:
results = act.discovery.download_data(ARM_USERNAME,
ARM_TOKEN,
'gucldM1.b1',
DATE,
DATE,
output=INSITU_DIR)
[DOWNLOADING] gucldM1.b1.20220314.000000.cdf
# Define the file path
nld = sorted(glob.glob(INSITU_DIR + '*gucldM1.b1.' + DATE.replace('-', '') + '*'))
# Define the site location based on the filename
ld_site = nld[0].split('gucld')[-1].split('.')[0]
# Call Match Datasets ACT
ds = match_datasets_act(ds, nld[0], ld_site, discard=discard_var['LD'])
4) Add Second Laser Disdrometer - S2 Site#
# We can use the ACT module for downloading data from the ARM web service
if ARM_USERNAME is None or ARM_TOKEN is None or len(ARM_USERNAME) == 0 or len(ARM_TOKEN) == 0:
print("ACT Username and Token Not Found ")
else:
results = act.discovery.download_data(ARM_USERNAME,
ARM_TOKEN,
'gucldS2.b1',
DATE,
DATE,
output=INSITU_DIR)
[DOWNLOADING] gucldS2.b1.20220314.000000.cdf
# Define the file path
ld_2 = sorted(glob.glob(INSITU_DIR + '*gucldS2.b1.' + DATE.replace('-', '') + '*'))
# Define the site location based on the filename
ld2_site = ld_2[0].split('gucld')[-1].split('.')[0]
# Call Match Datasets ACT
ds = match_datasets_act(ds, ld_2[0], ld2_site, discard=discard_var['LD'])
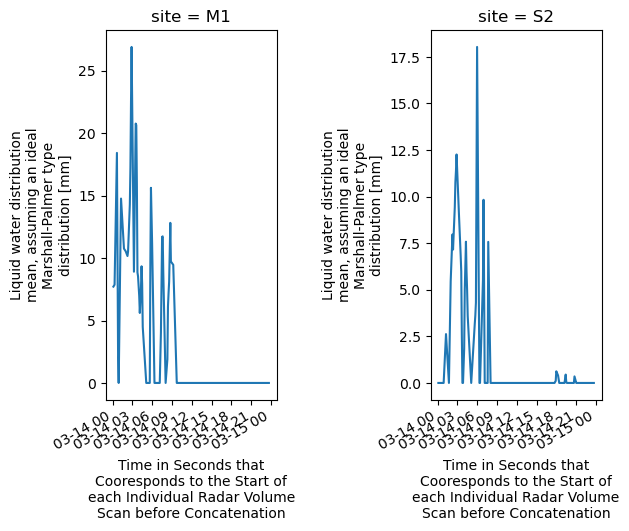
# Sanity Check - Plot LD from M1 and S2 sites to verify datastreams merged correctly
fig, axarr = plt.subplots(1, 2)
plt.subplots_adjust(wspace=0.9, hspace=1.2)
ds.liquid_water_distribution_mean.sel(site='M1').plot(ax=axarr[0])
ds.liquid_water_distribution_mean.sel(site='S2').plot(ax=axarr[1])
[<matplotlib.lines.Line2D at 0x1481e26b0>]
5) Add the Radar Wind Profiler (High Powered)#
NOTE: RWP Data are missing for March 14, 2022; ACT Discovery will alert if data are unavailable#
# We can use the ACT module for downloading data from the ARM web service
if ARM_USERNAME is None or ARM_TOKEN is None or len(ARM_USERNAME) == 0 or len(ARM_TOKEN) == 0:
print("ACT Username and Token Not Found ")
else:
results = act.discovery.download_data(ARM_USERNAME,
ARM_TOKEN,
'guc915rwpprecipmomenthighM1.a0',
DATE,
DATE,
output=INSITU_DIR)
No files returned or url status error.
Check datastream name, start, and end date.
# Define the file path
nrwp = sorted(glob.glob(INSITU_DIR + '*guc915rwpprecipmomenthighM1.a0.' + DATE.replace('-', '') + '*'))
if len(nrwp) > 0:
# Define the site location based on the filename
rwp_site = nrwp[0].split('guc915rwpprecipmomenthigh')[-1].split('.')[0]
# Call Match Datasets ACT
ds = match_datasets_act(ds, nrwp[0], rwp_site, resample='mean', discard=discard_var['RWP'])
# Check to make sure the RWP datastream merged in correctly
ds.sel(site='M1').doppler_velocity.plot(x='time')
6) Add the Radiosonde Data#
Note: There could be multiple launches per day and not all are successful.#
# We can use the ACT module for downloading data from the ARM web service
if ARM_USERNAME is None or ARM_TOKEN is None or len(ARM_USERNAME) == 0 or len(ARM_TOKEN) == 0:
print("ACT Username and Token Not Found ")
else:
results = act.discovery.download_data(ARM_USERNAME,
ARM_TOKEN,
'gucsondewnpnM1',
DATE,
DATE,
output=INSITU_DIR)
[DOWNLOADING] gucsondewnpnM1.b1.20220314.113200.cdf
[DOWNLOADING] gucsondewnpnM1.b1.20220314.115700.cdf
[DOWNLOADING] gucsondewnpnM1.b1.20220314.233000.cdf
# Search for all sonde files
nsonde = sorted(glob.glob(INSITU_DIR + "*gucsondewnpnM1.b1." + DATE.replace('-', '') + '*'))
# Open each individual Sonde File and Merge together
sonde_list = []
for nfile in nsonde:
sonde = act.io.armfiles.read_netcdf(nfile)
# Check to make sure launch was successful (e.g. time > 1 obs)
if sonde.time.shape[0] > 1:
sonde_list.append(sonde)
# Concatenate together the sonde xarray DataSets
ds_sonde = xr.concat(sonde_list, dim='time').compute()
# Define the site location based on the filename
sonde_site = nsonde[0].split('gucsondewnpn')[-1].split('.')[0]
ds = match_datasets_act(ds, ds_sonde, sonde_site, DataSet=True, discard=discard_var['Sonde'])
ds.sel(site='M1').alt.plot()
[<matplotlib.lines.Line2D at 0x153e5b130>]
7) Define the Meta Data Standards for Matched Dataset#
# Call the Data Object Definitions for this datastream.
# Will create an xarray dataset which will contain the necessary meta data and variables.
mdims = {"time": ds['time'].data.shape[0], "height": 70, "site": 8, "particle_size": 32, "raw_fall_velocity": 32}
out_ds = act.io.create_obj_from_arm_dod('xprecipradarradclss.c2', mdims, version='1.0')
# Transform the matched dataset for consistent dimensions
ds = ds.transpose('time', 'height', 'site', 'particle_size', 'raw_fall_velocity')
# Output Dataset has correct data attributes, supplied by the DOD.
# Update the output dataset variable values with the matched dataset.
for var in out_ds.variables:
if var not in out_ds.dims:
# check to see if variable is within the matched dataset
# note: it may not be if file is missing.
if var in ds.variables:
out_ds[var].data = ds[var].data
# Update the coordinates with the matched dataset values
out_ds = out_ds.assign_coords(time = ds['time'].data,
height = ds['height'].data,
site = ds['site'].data,
particle_size = ds['particle_size'].data,
raw_fall_velocity = ds['raw_fall_velocity'].data)
out_ds
<xarray.Dataset>
Dimensions: (time: 118, height: 70, site: 8,
particle_size: 32,
raw_fall_velocity: 32)
Coordinates:
* time (time) datetime64[ns] 2022-03-1...
* height (height) float64 3.149e+03 ... ...
* site (site) <U14 'M1' ... 'snodgrass'
* particle_size (particle_size) float32 0.062 ....
* raw_fall_velocity (raw_fall_velocity) float32 0.0...
Data variables: (12/121)
DBZ (time, height, site) float64 na...
VEL (time, height, site) float64 na...
WIDTH (time, height, site) float64 na...
ZDR (time, height, site) float64 na...
PHIDP (time, height, site) float64 na...
RHOHV (time, height, site) float64 na...
... ...
v_wind (time, site) float64 nan ... nan
wstat (time, site) float64 nan ... nan
asc (time, site) float64 nan ... nan
lat (site) float64 -9.999e+03 ... -...
lon (site) float64 -9.999e+03 ... -...
alt (time, site) float64 nan ... nan
Attributes: (12/24)
command_line:
Conventions: ARM-1.3 CF/Radial instrument_parameters
process_version:
dod_version:
input_datastreams:
site_id:
... ...
developers: Joseph O'Brien, ANL. Maxwell Grover, ANL. Robert J...
translator: https://www.arm.gov/capabilities/instruments/xprec...
mentors: https://www.arm.gov/connect-with-arm/organization/...
source: Colorado State University's X-Band Precipitation R...
field_names: DBZ, VEL, WIDTH, ZDR, PHIDP, RHOHV, NCP, DBZhv, cb...
history: xarray.Dataset
- time: 118
- height: 70
- site: 8
- particle_size: 32
- raw_fall_velocity: 32
- time(time)datetime64[ns]2022-03-14T00:06:47 ... 2022-03-...
array(['2022-03-14T00:06:47.000000000', '2022-03-14T00:17:28.000000000', '2022-03-14T00:38:47.000000000', '2022-03-14T00:49:28.000000000', '2022-03-14T00:54:48.000000000', '2022-03-14T01:16:08.000000000', '2022-03-14T01:42:48.000000000', '2022-03-14T01:58:48.000000000', '2022-03-14T02:14:48.000000000', '2022-03-14T02:20:08.000000000', '2022-03-14T02:36:08.000000000', '2022-03-14T02:41:28.000000000', '2022-03-14T02:46:48.000000000', '2022-03-14T02:52:08.000000000', '2022-03-14T03:13:29.000000000', '2022-03-14T03:29:28.000000000', '2022-03-14T03:34:49.000000000', '2022-03-14T03:40:09.000000000', '2022-03-14T03:45:29.000000000', '2022-03-14T03:50:49.000000000', '2022-03-14T04:01:28.000000000', '2022-03-14T04:06:49.000000000', '2022-03-14T04:17:29.000000000', '2022-03-14T04:22:49.000000000', '2022-03-14T04:28:09.000000000', '2022-03-14T04:33:29.000000000', '2022-03-14T05:05:29.000000000', '2022-03-14T05:37:29.000000000', '2022-03-14T05:42:49.000000000', '2022-03-14T05:48:09.000000000', '2022-03-14T05:58:49.000000000', '2022-03-14T06:20:10.000000000', '2022-03-14T06:25:29.000000000', '2022-03-14T06:52:10.000000000', '2022-03-14T06:57:30.000000000', '2022-03-14T07:08:10.000000000', '2022-03-14T07:18:50.000000000', '2022-03-14T07:29:30.000000000', '2022-03-14T07:34:50.000000000', '2022-03-14T07:40:10.000000000', '2022-03-14T08:01:30.000000000', '2022-03-14T08:17:30.000000000', '2022-03-14T08:22:50.000000000', '2022-03-14T08:28:10.000000000', '2022-03-14T08:33:30.000000000', '2022-03-14T08:44:10.000000000', '2022-03-14T08:49:30.000000000', '2022-03-14T09:10:51.000000000', '2022-03-14T09:42:50.000000000', '2022-03-14T09:53:30.000000000', '2022-03-14T09:58:51.000000000', '2022-03-14T10:20:11.000000000', '2022-03-14T10:25:31.000000000', '2022-03-14T10:41:31.000000000', '2022-03-14T11:24:11.000000000', '2022-03-14T11:29:31.000000000', '2022-03-14T11:40:11.000000000', '2022-03-14T12:01:31.000000000', '2022-03-14T12:06:52.000000000', '2022-03-14T12:12:11.000000000', '2022-03-14T12:38:51.000000000', '2022-03-14T12:44:11.000000000', '2022-03-14T12:54:52.000000000', '2022-03-14T13:00:12.000000000', '2022-03-14T13:05:32.000000000', '2022-03-14T13:16:12.000000000', '2022-03-14T13:21:32.000000000', '2022-03-14T13:26:52.000000000', '2022-03-14T13:53:32.000000000', '2022-03-14T14:04:13.000000000', '2022-03-14T14:25:32.000000000', '2022-03-14T15:34:52.000000000', '2022-03-14T15:40:13.000000000', '2022-03-14T15:50:53.000000000', '2022-03-14T16:01:33.000000000', '2022-03-14T16:22:53.000000000', '2022-03-14T16:38:53.000000000', '2022-03-14T16:49:33.000000000', '2022-03-14T16:54:53.000000000', '2022-03-14T17:05:33.000000000', '2022-03-14T17:16:13.000000000', '2022-03-14T17:26:53.000000000', '2022-03-14T17:37:33.000000000', '2022-03-14T17:42:53.000000000', '2022-03-14T17:53:34.000000000', '2022-03-14T17:58:53.000000000', '2022-03-14T18:14:54.000000000', '2022-03-14T18:25:34.000000000', '2022-03-14T18:30:54.000000000', '2022-03-14T18:46:54.000000000', '2022-03-14T18:57:34.000000000', '2022-03-14T19:02:53.000000000', '2022-03-14T19:08:14.000000000', '2022-03-14T19:13:34.000000000', '2022-03-14T19:24:14.000000000', '2022-03-14T19:29:34.000000000', '2022-03-14T19:34:54.000000000', '2022-03-14T19:56:14.000000000', '2022-03-14T20:01:34.000000000', '2022-03-14T20:12:14.000000000', '2022-03-14T20:17:34.000000000', '2022-03-14T20:38:54.000000000', '2022-03-14T20:44:14.000000000', '2022-03-14T21:00:15.000000000', '2022-03-14T21:10:54.000000000', '2022-03-14T21:21:34.000000000', '2022-03-14T22:04:15.000000000', '2022-03-14T22:09:35.000000000', '2022-03-14T22:25:35.000000000', '2022-03-14T22:36:15.000000000', '2022-03-14T22:41:35.000000000', '2022-03-14T22:52:15.000000000', '2022-03-14T23:02:55.000000000', '2022-03-14T23:13:35.000000000', '2022-03-14T23:24:15.000000000', '2022-03-14T23:29:35.000000000', '2022-03-14T23:40:16.000000000', '2022-03-15T00:01:36.000000000'], dtype='datetime64[ns]') - height(height)float643.149e+03 3.249e+03 ... 1.005e+04
array([ 3149., 3249., 3349., 3449., 3549., 3649., 3749., 3849., 3949., 4049., 4149., 4249., 4349., 4449., 4549., 4649., 4749., 4849., 4949., 5049., 5149., 5249., 5349., 5449., 5549., 5649., 5749., 5849., 5949., 6049., 6149., 6249., 6349., 6449., 6549., 6649., 6749., 6849., 6949., 7049., 7149., 7249., 7349., 7449., 7549., 7649., 7749., 7849., 7949., 8049., 8149., 8249., 8349., 8449., 8549., 8649., 8749., 8849., 8949., 9049., 9149., 9249., 9349., 9449., 9549., 9649., 9749., 9849., 9949., 10049.]) - site(site)<U14'M1' 'S2' ... 'snodgrass'
array(['M1', 'S2', 'avery_point', 'brush_creek', 'kettle_ponds', 'pumphouse_site', 'roaring_judy', 'snodgrass'], dtype='<U14') - particle_size(particle_size)float320.062 0.187 0.312 ... 21.5 24.0
array([ 0.062, 0.187, 0.312, 0.437, 0.562, 0.687, 0.812, 0.937, 1.062, 1.187, 1.375, 1.625, 1.875, 2.125, 2.375, 2.75 , 3.25 , 3.75 , 4.25 , 4.75 , 5.5 , 6.5 , 7.5 , 8.5 , 9.5 , 11. , 13. , 15. , 17. , 19. , 21.5 , 24. ], dtype=float32) - raw_fall_velocity(raw_fall_velocity)float320.05 0.15 0.25 ... 15.2 17.6 20.8
array([ 0.05, 0.15, 0.25, 0.35, 0.45, 0.55, 0.65, 0.75, 0.85, 0.95, 1.1 , 1.3 , 1.5 , 1.7 , 1.9 , 2.2 , 2.6 , 3. , 3.4 , 3.8 , 4.4 , 5.2 , 6. , 6.8 , 7.6 , 8.8 , 10.4 , 12. , 13.6 , 15.2 , 17.6 , 20.8 ], dtype=float32)
- DBZ(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Equaivalent Radar Reflectivity Factor
- units :
- dBZ
- _FillValue :
- -999999.0
- standard_name :
- equivalent_reflectivity_factor
- coordinates :
- elevation azimuth range
- source :
- xprecipradarcmacppi.c1
array([[[ nan, nan, nan, ..., nan, nan, nan], [-10.81999969, nan, nan, ..., 7.30825315, nan, -10.93107905], [ -6.31648953, nan, 14.72636585, ..., 4.06749179, nan, -4.84339817], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [-10.81999969, nan, nan, ..., -2.96758446, nan, 7.9337979 ], [ -5.41896141, nan, 17.84486603, ..., 15.24007766, nan, 20.81337317], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [-10.81999969, nan, nan, ..., -15.92000008, nan, 7.10398186], [-10.81999969, nan, -11.6936225 , ..., -15.92000008, nan, -11.84000015], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - VEL(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Radial Doppler Velocity, Positive for Motion Away from Instrument
- units :
- m/s
- _FillValue :
- -999999.0
- standard_name :
- radial_velocity_of_scatterers_away_from_instruments
- coordinates :
- elevation azimuth range
- source :
- xprecipradarcmacppi.c1
array([[[ nan, nan, nan, ..., nan, nan, nan], [ 12.40704339, nan, nan, ..., -2.02765943, nan, -22.50603496], [-10.28064785, nan, -1.03943419, ..., -1.73831865, nan, -3.05127251], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ 12.16807962, nan, nan, ..., -30.57713021, nan, 12.46009315], [ -9.74842282, nan, 0.06915687, ..., 2.47852138, nan, -2.41579192], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ 11.12294777, nan, nan, ..., 21.93758999, nan, -26.08425652], [ -6.95099764, nan, 14.87201837, ..., -19.01108051, nan, -3.89201876], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - WIDTH(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Spectral Width
- units :
- m/s
- _FillValue :
- -999999.0
- standard_name :
- doppler_spectrum_width
- coordinates :
- elevation azimuth range
- source :
- xprecipradarcmacppi.c1
array([[[ nan, nan, nan, ..., nan, nan, nan], [-9.99000000e+04, nan, nan, ..., 7.58765939e-01, nan, -9.99000000e+04], [-8.22842531e+04, nan, 1.03685529e+00, ..., 7.29957409e-01, nan, -9.99000000e+04], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [-9.99000000e+04, nan, nan, ..., -9.37358587e+04, nan, -9.99000000e+04], [-8.22843782e+04, nan, 1.05535844e+00, ..., -1.25400256e+04, nan, -6.82427587e+04], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [-9.99000000e+04, nan, nan, ..., 2.84406397e+00, nan, -9.99000000e+04], [-9.99000000e+04, nan, -7.86631615e+04, ..., 3.18489355e+00, nan, -9.99000000e+04], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - ZDR(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Differential Reflectivity
- units :
- dB
- _FillValue :
- -999999.0
- standard_name :
- log_differential_reflectivity_hv
- coordinates :
- elevation azimuth range
- source :
- xprecipradarcmacppi.c1
array([[[ nan, nan, nan, ..., nan, nan, nan], [ 1.72313261e+00, nan, nan, ..., 2.30185108e+00, nan, 7.39442602e+00], [ 1.94521916e+00, nan, 2.07116997e+00, ..., 2.34631925e+00, nan, 1.33644228e+00], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [-1.54023688e+04, nan, nan, ..., 2.31617026e+00, nan, 4.99945852e+00], [-8.22815469e+04, nan, 2.31062895e+00, ..., 2.46900026e+00, nan, 3.72607752e+00], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [-1.54012609e+04, nan, nan, ..., 2.72776620e+00, nan, -6.91175989e+04], [-9.98969500e+04, nan, 8.27885439e-01, ..., 3.14497870e+00, nan, 1.64093429e+00], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - PHIDP(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Differential Phase
- units :
- degree
- _FillValue :
- -999999.0
- standard_name :
- differential_phase_hv
- coordinates :
- elevation azimuth range
- source :
- xprecipradarcmacppi.c1
array([[[ nan, nan, nan, ..., nan, nan, nan], [128.32599258, nan, nan, ..., 100.52865903, nan, 233.82960237], [ 66.94012996, nan, 99.0469331 , ..., 100.0267886 , nan, 101.40374094], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [102.38336946, nan, nan, ..., 99.92305943, nan, 118.80084311], [124.70658576, nan, 101.87880195, ..., 100.6697407 , nan, 97.36773662], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ 79.69283065, nan, nan, ..., 292.30180737, nan, 85.45987434], [140.7436992 , nan, 271.93664734, ..., 231.35807351, nan, 203.35780229], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - RHOHV(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Cross-Polar Correlation Ratio
- units :
- 1
- _FillValue :
- -999999.0
- standard_name :
- cross_correlation_ratio_hv
- coordinates :
- elevation azimuth range
- source :
- xprecipradarcmacppi.c1
array([[[ nan, nan, nan, ..., nan, nan, nan], [ 8.44771128e-01, nan, nan, ..., 9.95876644e-01, nan, 6.14005694e-01], [ 8.73862805e-01, nan, 9.89913246e-01, ..., 9.94376618e-01, nan, 9.13857231e-01], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ 9.40221613e-01, nan, nan, ..., 9.86740491e-01, nan, 7.98946131e-01], [ 7.88435599e-01, nan, 9.91062963e-01, ..., 9.98000026e-01, nan, 9.72922131e-01], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ 9.84233782e-01, nan, nan, ..., 5.86753201e-01, nan, 7.96713342e-01], [ 9.66666287e-01, nan, 6.46880552e-01, ..., 6.05972443e-01, nan, 5.25898647e-01], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - NCP(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Normalized Coherent Power, also known as SQI
- units :
- 1
- _FillValue :
- -999999.0
- standard_name :
- normalized_coherent_power
- coordinates :
- elevation azimuth range
- source :
- xprecipradarcmacppi.c1
array([[[ nan, nan, nan, ..., nan, nan, nan], [0.96999997, nan, nan, ..., 0.95999998, nan, 0.96999997], [0.96118338, nan, 0.93425158, ..., 0.9612553 , nan, 0.96683114], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [0.96999997, nan, nan, ..., 0.95999998, nan, 0.95151344], [0.96999997, nan, 0.94425157, ..., 0.9612553 , nan, 0.9036623 ], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [0.96999997, nan, nan, ..., 0.11 , nan, 0.96999997], [0.93826027, nan, 0.77961 , ..., 0.1049787 , nan, 0.96674664], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - DBZhv(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Equivalent Reflectivity Factor HV
- units :
- dBZ
- _FillValue :
- -999999.0
- standard_name :
- equivalent_reflectivity_factor_hv
- coordinates :
- elevation azimuth range
- source :
- xprecipradarcmacppi.c1
array([[[ nan, nan, nan, ..., nan, nan, nan], [ 5.76836845e+00, nan, nan, ..., 7.66961466e+00, nan, 4.88642299e+00], [ 1.58196087e+01, nan, 1.43160390e+01, ..., 4.31130019e+00, nan, -1.81306186e+00], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ 9.12104147e+00, nan, nan, ..., 1.06975213e+00, nan, 1.80643284e+01], [ 1.74641430e+01, nan, 1.78260997e+01, ..., 1.42095936e+01, nan, 2.02568279e+01], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ 9.74759839e+00, nan, nan, ..., -2.85639758e+01, nan, 1.58022412e+01], [ 1.57818002e+01, nan, -7.42380933e+00, ..., -2.36445975e+01, nan, -1.66800558e+01], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - cbb_flag(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Cumulative Beam Block Fraction Flag
- units :
- 1
- _FillValue :
- -999999.0
- coordinates :
- elevation azimuth range
- source :
- xprecipradarcmacppi.c1
array([[[ nan, nan, nan, ..., nan, nan, nan], [0.84578254, nan, nan, ..., 0. , nan, 0. ], [0. , nan, 0. , ..., 0. , nan, 0. ], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [0.84578254, nan, nan, ..., 0. , nan, 0. ], [0. , nan, 0. , ..., 0. , nan, 0. ], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [0.84578254, nan, nan, ..., 0. , nan, 0. ], [0. , nan, 0. , ..., 0. , nan, 0. ], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - sounding_temperature(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Interpolated profile
- units :
- degC
- _FillValue :
- -999999.0
- standard_name :
- interpolated_profile
- source :
- xprecipradarcmacppi.c1
array([[[ nan, nan, nan, ..., nan, nan, nan], [-3.02489149, nan, nan, ..., -3.0108487 , nan, -3.03686548], [-4.00664871, nan, -3.99916278, ..., -4.00585198, nan, -4.00033004], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [-3.02489149, nan, nan, ..., -3.0108487 , nan, -3.03686548], [-4.00664871, nan, -3.99916278, ..., -4.00585198, nan, -4.00033004], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [-3.25761667, nan, nan, ..., -3.2624767 , nan, -3.24478044], [-4.22537902, nan, -4.22329579, ..., -4.20459192, nan, -4.21064839], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - signal_to_noise_ratio(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Signal to Noise Ratio
- units :
- dB
- _FillValue :
- -999999.0
- standard_name :
- signal_to_noise_ratio
- coordinates :
- elevation azimuth range
- source :
- xprecipradarcmacppi.c1
array([[[ nan, nan, nan, ..., nan, nan, nan], [ 0. , nan, nan, ..., 23.23763593, nan, 0.90892088], [ 4.50351016, nan, 20.02361069, ..., 19.9887468 , nan, 6.9617442 ], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ 0. , nan, nan, ..., 12.94303243, nan, 19.78379846], [ 5.40280164, nan, 23.14211258, ..., 31.161333 , nan, 32.62534888], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ 0. , nan, nan, ..., 0. , nan, 18.95090042], [ 0. , nan, -6.39637728, ..., 0. , nan, 0. ], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - velocity_texture(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Mean dopper velocity
- units :
- m/s
- _FillValue :
- -999999.0
- standard_name :
- radial_velocity_of_scatterers_away_from_instrument
- coordinates :
- elevation azimuth range
- source :
- xprecipradarcmacppi.c1
array([[[ nan, nan, nan, ..., nan, nan, nan], [4.34760833, nan, nan, ..., 0.6256861 , nan, 4.18866136], [2.36289549, nan, 1.61258041, ..., 0.64153505, nan, 3.44449815], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [4.24774887, nan, nan, ..., 5.31099301, nan, 3.28332395], [2.32391028, nan, 2.28895862, ..., 5.90383218, nan, 0.52435537], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [4.33351278, nan, nan, ..., 5.62090783, nan, 4.52300875], [5.17077991, nan, 6.73562328, ..., 6.32464634, nan, 5.60383135], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - gate_id(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Classification of dominant scatterer
- units :
- 1
- notes :
- 0:multi_trip,1:rain,2:snow,3:no_scatter,4:melting,5:clutter,6:terrain_blockage
- valid_max :
- 6
- valid_min :
- 0
- flag_values :
- 0, 1, 2, 3, 4, 5, 6
- flag_meanings :
- multi_trip rain snow no_scatter melting clutter terrain_blockage
- source :
- xprecipradarcmacppi.c1
array([[[ nan, nan, nan, ..., nan, nan, nan], [5.53734763, nan, nan, ..., 1.87659471, nan, 3. ], [2.82366835, nan, 2. , ..., 0.25106487, nan, 2.68311657], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [5.53734763, nan, nan, ..., 0. , nan, 0.61621754], [2.82366835, nan, 0.42515785, ..., 0. , nan, 2. ], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [5.53734763, nan, nan, ..., 3. , nan, 3. ], [3. , nan, 3. , ..., 3. , nan, 3. ], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - simulated_velocity(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Simulated mean doppler velocity
- units :
- m/s
- _FillValue :
- -999999.0
- standard_name :
- radial_velocity_of_scatterers_away_from_instrument
- coordinates :
- elevation azimuth range
- source :
- xprecipradarcmacppi.c1
array([[[ nan, nan, nan, ..., nan, nan, nan], [-3.51907816, nan, nan, ..., -3.90425293, nan, -3.68023146], [-2.87505424, nan, -3.17556306, ..., -3.32425956, nan, -2.94900991], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [-3.52440266, nan, nan, ..., -3.90299974, nan, -3.67899042], [-2.87806457, nan, -3.1751076 , ..., -3.32474269, nan, -2.95073851], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [-5.74909365, nan, nan, ..., -5.39203247, nan, -5.94086844], [-5.65848637, nan, -5.91648484, ..., -5.59354396, nan, -5.81614524], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - corrected_velocity(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Corrected mean doppler velocity
- units :
- m/s
- _FillValue :
- -999999.0
- standard_name :
- corrected_radial_velocity_of_scatterers_away_from_instrument
- valid_max :
- 47.7
- valid_min :
- -47.7
- coordinates :
- elevation azimuth range
- source :
- xprecipradarcmacppi.c1
array([[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, -1.03943419, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, -2.41579192], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - unfolded_differential_phase(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Unfolded differential propagation phase shift
- units :
- degree
- _FillValue :
- -999999.0
- standard_name :
- differential_phase_hv
- coordinates :
- elevation azimuth range
- source :
- xprecipradarcmacppi.c1
array([[[nan, nan, nan, ..., nan, nan, nan], [ 0., nan, nan, ..., 0., nan, 0.], [ 0., nan, 0., ..., 0., nan, 0.], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [ 0., nan, nan, ..., 0., nan, 0.], [ 0., nan, 0., ..., 0., nan, 0.], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [ 0., nan, nan, ..., 0., nan, 0.], [ 0., nan, 0., ..., 0., nan, 0.], ..., ... ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [ 0., nan, nan, ..., 0., nan, 0.], [ 0., nan, 0., ..., 0., nan, 0.], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [ 0., nan, nan, ..., 0., nan, 0.], [ 0., nan, 0., ..., 0., nan, 0.], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]]) - corrected_differential_phase(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Corrected differential propagation phase shift
- units :
- degree
- _FillValue :
- -999999.0
- standard_name :
- differential_phase_hv
- valid_max :
- 400.0
- valid_min :
- 0.0
- coordinates :
- elevation azimuth range
- source :
- xprecipradarcmacppi.c1
array([[[ nan, nan, nan, ..., nan, nan, nan], [0.01, nan, nan, ..., 0.01, nan, 0.01], [0.01, nan, 0.01, ..., 0.01, nan, 0.01], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [0.01, nan, nan, ..., 0.01, nan, 0.01], [0.01, nan, 0.01, ..., 0.01, nan, 0.01], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [0.01, nan, nan, ..., 0.01, nan, 0.01], [0.01, nan, 0.01, ..., 0.01, nan, 0.01], ..., ... ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [0.01, nan, nan, ..., 0.01, nan, 0.01], [0.01, nan, 0.01, ..., 0.01, nan, 0.01], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [0.01, nan, nan, ..., 0.01, nan, 0.01], [0.01, nan, 0.01, ..., 0.01, nan, 0.01], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - filtered_corrected_differential_phase(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Filtered Corrected Differential Phase
- units :
- degree
- _FillValue :
- -999999.0
- standard_name :
- differential_phase_hv
- valid_max :
- 400.0
- valid_min :
- 0.0
- coordinates :
- elevation azimuth range
- source :
- xprecipradarcmacppi.c1
array([[[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., ... ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [ 0., nan, nan, ..., 0., nan, 0.], [ 0., nan, 0., ..., 0., nan, 0.], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [ 0., nan, nan, ..., 0., nan, 0.], [ 0., nan, 0., ..., 0., nan, 0.], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]]) - corrected_specific_diff_phase(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Specific differential phase (KDP)
- units :
- degrees/km
- _FillValue :
- -999999.0
- standard_name :
- specific_differential_phase_hv
- coordinates :
- elevation azimuth range
- source :
- xprecipradarcmacppi.c1
array([[[nan, nan, nan, ..., nan, nan, nan], [ 0., nan, nan, ..., 0., nan, 0.], [ 0., nan, 0., ..., 0., nan, 0.], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [ 0., nan, nan, ..., 0., nan, 0.], [ 0., nan, 0., ..., 0., nan, 0.], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [ 0., nan, nan, ..., 0., nan, 0.], [ 0., nan, 0., ..., 0., nan, 0.], ..., ... ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [ 0., nan, nan, ..., 0., nan, 0.], [ 0., nan, 0., ..., 0., nan, 0.], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [ 0., nan, nan, ..., 0., nan, 0.], [ 0., nan, 0., ..., 0., nan, 0.], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]]) - filtered_corrected_specific_diff_phase(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Filtered Corrected Specific differential phase (KDP)
- units :
- degrees/km
- _FillValue :
- -999999.0
- standard_name :
- specific_differential_phase_hv
- coordinates :
- elevation azimuth range
- source :
- xprecipradarcmacppi.c1
array([[[nan, nan, nan, ..., nan, nan, nan], [ 0., nan, nan, ..., 0., nan, 0.], [ 0., nan, 0., ..., 0., nan, 0.], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [ 0., nan, nan, ..., 0., nan, 0.], [ 0., nan, 0., ..., 0., nan, 0.], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [ 0., nan, nan, ..., 0., nan, 0.], [ 0., nan, 0., ..., 0., nan, 0.], ..., ... ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [ 0., nan, nan, ..., 0., nan, 0.], [ 0., nan, 0., ..., 0., nan, 0.], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [ 0., nan, nan, ..., 0., nan, 0.], [ 0., nan, 0., ..., 0., nan, 0.], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]]) - corrected_differential_reflectivity(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Corrected differential reflectivity
- units :
- dB
- _FillValue :
- -999999.0
- standard_name :
- corrected_log_differential_reflectivity_hv
- coordinates :
- elevation azimuth range
- source :
- xprecipradarcmacppi.c1
array([[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, 2.07116997, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, 3.72607752], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - corrected_reflectivity(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Corrected reflectivity
- units :
- dBZ
- _FillValue :
- -999999.0
- standard_name :
- corrected_equivalent_reflectivity_factor
- coordinates :
- elevation azimuth range
- source :
- xprecipradarcmacppi.c1
array([[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, 14.72636585, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, 20.81337317], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - height_over_iso0(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Height of radar beam over freezing level
- units :
- m
- _FillValue :
- -999999.0
- standard_name :
- height
- source :
- xprecipradarcmacppi.c1
array([[[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., ... ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]]) - specific_attenuation(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Specific attenuation
- units :
- dB/km
- _FillValue :
- -999999.0
- standard_name :
- specific_attenuation
- valid_max :
- 1.0
- valid_min :
- 0.0
- coordinates :
- elevation azimuth range
- source :
- xprecipradarcmacppi.c1
array([[[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, 0., ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, 0.], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, 0.], ..., ... ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]]) - path_integrated_attenuation(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Path Integrated Attenuation
- units :
- dB
- _FillValue :
- -999999.0
- coordinates :
- elevation azimuth range
- source :
- xprecipradarcmacppi.c1
array([[[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, 0., ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, 0.], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, 0.], ..., ... ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]]) - specific_differential_attenuation(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Specific Differential Attenuation
- units :
- dB/km
- _FillValue :
- -999999.0
- coordinates :
- elevation azimuth range
- source :
- xprecipradarcmacppi.c1
array([[[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, 0., ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, 0.], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, 0.], ..., ... ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]]) - path_integrated_differential_attenuation(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Path Integrated Differential Attenuation
- units :
- dB
- _FillValue :
- -999999.0
- coordinates :
- elevation azimuth range
- source :
- xprecipradarcmacppi.c1
array([[[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, 0., ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, 0.], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, 0.], ..., ... ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]]) - rain_rate_A(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Rainfall Rate from Specific Attenuation
- units :
- mm/hr
- _FillValue :
- -999999.0
- standard_name :
- rainfall_rate
- valid_max :
- 400.0
- valid_min :
- 0.0
- coordinates :
- elevation azimuth range
- source :
- xprecipradarcmacppi.c1
- comment :
- Rain rate calculated from specific_attenuation, R=43.5*specific_attenuation**0.79, note R=0.0 where norm coherent power < 0.4 or rhohv < 0.8
array([[[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, 0., ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, 0.], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, 0.], ..., ... ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]]) - snow_rate_ws2012(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Snowfall rate from Z using Wolf and Snider (2012)
- units :
- mm/h
- _FillValue :
- -999999.0
- standard_name :
- snowfall_rate
- valid_max :
- 500
- valid_min :
- 0
- coordinates :
- elevation azimuth range
- source :
- xprecipradarcmacppi.c1
array([[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, 7.3066064 , ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, 15.40165278], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - snow_rate_ws88diw(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Snowfall rate from Z using WSR 88D High Plains
- units :
- mm/h
- _FillValue :
- -999999.0
- standard_name :
- snowfall_rate
- valid_max :
- 500
- valid_min :
- 0
- coordinates :
- elevation azimuth range
- source :
- xprecipradarcmacppi.c1
array([[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, 12.11663596, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, 25.54075172], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - snow_rate_m2009_1(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Snowfall rate from Z using Matrosov et al.(2009) Braham(1990) 1
- units :
- mm/h
- _FillValue :
- -999999.0
- standard_name :
- snowfall_rate
- valid_max :
- 500
- valid_min :
- 0
- coordinates :
- elevation azimuth range
- source :
- xprecipradarcmacppi.c1
array([[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, 7.75818776, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, 25.44635936], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - snow_rate_m2009_2(time, height, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Snowfall rate from Z using Matrosov et al.(2009) Braham(1990) 2
- units :
- mm/h
- _FillValue :
- -999999.0
- standard_name :
- snowfall_rate
- valid_max :
- 500
- valid_min :
- 0
- coordinates :
- elevation azimuth range
- source :
- xprecipradarcmacppi.c1
array([[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, 5.50638925, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, 16.37988589], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - intensity_rt(time, site)float640.0 nan nan nan ... nan nan nan nan
- long_name :
- Heavy precipitation alarm
- units :
- mm/hr
- _FillValue :
- -999999.0
- valid_min :
- 0.0
- valid_max :
- 3000.0
- threshold :
- 6 mm/hr
- absolute_accuracy :
- plus/minus 6
- comment_1 :
- Only measurements that exceed the threshold are recorded. Any measurement below the threshold is reported as 0 mm/hr.
- source :
- gucwbpluvio2M1.a1
array([[ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], ... [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan]]) - accum_rtnrt(time, site)float640.0 nan nan nan ... nan nan nan nan
- long_name :
- Accumulated amounts of precipitation over the sampling interval exceeding a threshold of 0.05mm or the accumulated amount of fine precipitation observed over the last hour
- units :
- mm
- _FillValue :
- -999999.0
- valid_min :
- 0.0
- valid_max :
- 500.0
- threshold :
- 0.05 mm
- absolute_accuracy :
- plus/minus 0.1
- equation :
- The accum_rtnrt variable is calculated by first measuring the accumulated amount of rain in the last minute. If this measurement exceeds the threshold, it reports this real time value. If the real time measurement does not reach the threshold, it reports the non real time measurement using the same equation as the accum_nrt variable.
- comment :
- Only measurements that exceed the threshold are recorded. Any measurement below the threshold is reported as 0 mm.
- source :
- gucwbpluvio2M1.a1
array([[0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0.1288 , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0.2112 , nan, nan, nan, nan, nan, nan, nan], [0.06186667, nan, nan, nan, nan, nan, nan, nan], ... [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - accum_nrt(time, site)float640.0 nan nan nan ... nan nan nan nan
- long_name :
- Accumulated precipitation over the sampling interval filtered and delayed by 5 minute
- units :
- mm
- _FillValue :
- -999999.0
- valid_min :
- 0.0
- valid_max :
- 500.0
- threshold :
- 0.05 mm
- absolute_accuracy :
- plus/minus 0.1
- equation :
- The accum_nrt variable is calculated by measuring the amount of rain accumulate in a sampling interval at most 1 hour long, with the end of the interval at the given time. The start of the sampling interval occurs within the past hour, but is unknown. The start of the interval is determined once the accumulated sum either exceeds 0.05 or the interval length reaches an hour
- comment :
- Only measurements that exceed the threshold are recorded. Any measurement below the threshold is reported as 0 mm.
- source :
- gucwbpluvio2M1.a1
array([[0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0.1288 , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0.2208 , nan, nan, nan, nan, nan, nan, nan], [0.0524 , nan, nan, nan, nan, nan, nan, nan], ... [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - accum_total_nrt(time, site)float64252.5 nan nan nan ... nan nan nan
- long_name :
- Sum of accum_nrt values since the last device start
- units :
- mm
- _FillValue :
- -999999.0
- valid_min :
- 0.0
- valid_max :
- 500.0
- threshold :
- 0.05 mm
- absolute_accuracy :
- plus/minus 0.1
- comment :
- Only measurements that exceed the threshold are recorded. Any measurement below the threshold is reported as 0 mm.
- source :
- gucwbpluvio2M1.a1
array([[252.45001221, nan, nan, nan, nan, nan, nan, nan], [252.45001221, nan, nan, nan, nan, nan, nan, nan], [252.45001221, nan, nan, nan, nan, nan, nan, nan], [252.45001221, nan, nan, nan, nan, nan, nan, nan], [252.45001221, nan, nan, nan, nan, nan, nan, nan], [252.45001221, nan, nan, nan, nan, nan, nan, nan], [254.98160889, nan, nan, nan, nan, nan, nan, nan], [256. , nan, nan, nan, nan, nan, nan, nan], [256.22081055, nan, nan, nan, nan, nan, nan, nan], [257.30440796, nan, nan, nan, nan, nan, nan, nan], ... [280.30001831, nan, nan, nan, nan, nan, nan, nan], [280.30001831, nan, nan, nan, nan, nan, nan, nan], [280.30001831, nan, nan, nan, nan, nan, nan, nan], [280.30001831, nan, nan, nan, nan, nan, nan, nan], [280.30001831, nan, nan, nan, nan, nan, nan, nan], [280.30001831, nan, nan, nan, nan, nan, nan, nan], [280.30001831, nan, nan, nan, nan, nan, nan, nan], [280.30001831, nan, nan, nan, nan, nan, nan, nan], [280.30001831, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - bucket_rt(time, site)float641.61e+03 nan nan ... nan nan nan
- long_name :
- The currently measured, unfiltered bucket contents since last reset
- units :
- mm
- _FillValue :
- -999999.0
- valid_min :
- 20.0
- valid_max :
- 1800.0
- threshold :
- 0.01 mm
- absolute_accuracy :
- plus/minus 0.1
- comment :
- Only increases that exceed the threshold are recorded. Any increase less than threshold is reported as no increase
- source :
- gucwbpluvio2M1.a1
array([[1609.5 , nan, nan, nan, nan, nan, nan, nan], [1609.59864258, nan, nan, nan, nan, nan, nan, nan], [1610. , nan, nan, nan, nan, nan, nan, nan], [1610.5 , nan, nan, nan, nan, nan, nan, nan], [1610.5 , nan, nan, nan, nan, nan, nan, nan], [1611. , nan, nan, nan, nan, nan, nan, nan], [1613.22408203, nan, nan, nan, nan, nan, nan, nan], [1613.90402344, nan, nan, nan, nan, nan, nan, nan], [1614.78804687, nan, nan, nan, nan, nan, nan, nan], [1615.22128581, nan, nan, nan, nan, nan, nan, nan], ... [1638. , nan, nan, nan, nan, nan, nan, nan], [1638. , nan, nan, nan, nan, nan, nan, nan], [1638. , nan, nan, nan, nan, nan, nan, nan], [1638. , nan, nan, nan, nan, nan, nan, nan], [1638. , nan, nan, nan, nan, nan, nan, nan], [1638. , nan, nan, nan, nan, nan, nan, nan], [1638. , nan, nan, nan, nan, nan, nan, nan], [1638. , nan, nan, nan, nan, nan, nan, nan], [1638. , nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - bucket_nrt(time, site)float641.61e+03 nan nan ... nan nan nan
- long_name :
- The currently measured, filtered bucket contents since last reset
- units :
- mm
- _FillValue :
- -999999.0
- valid_min :
- 20.0
- valid_max :
- 1800.0
- threshold :
- 0.01 mm
- absolute_accuracy :
- plus/minus 0.1
- comment :
- Only increases that exceed the threshold are recorded. Any increase less than threshold is reported as no increase
- source :
- gucwbpluvio2M1.a1
array([[1609.5 , nan, nan, nan, nan, nan, nan, nan], [1609.5 , nan, nan, nan, nan, nan, nan, nan], [1610. , nan, nan, nan, nan, nan, nan, nan], [1610.45733073, nan, nan, nan, nan, nan, nan, nan], [1610.5 , nan, nan, nan, nan, nan, nan, nan], [1610.69074219, nan, nan, nan, nan, nan, nan, nan], [1612.82398926, nan, nan, nan, nan, nan, nan, nan], [1613.65196289, nan, nan, nan, nan, nan, nan, nan], [1614.49200195, nan, nan, nan, nan, nan, nan, nan], [1614.91068848, nan, nan, nan, nan, nan, nan, nan], ... [1638. , nan, nan, nan, nan, nan, nan, nan], [1638. , nan, nan, nan, nan, nan, nan, nan], [1638. , nan, nan, nan, nan, nan, nan, nan], [1638. , nan, nan, nan, nan, nan, nan, nan], [1638. , nan, nan, nan, nan, nan, nan, nan], [1638. , nan, nan, nan, nan, nan, nan, nan], [1638. , nan, nan, nan, nan, nan, nan, nan], [1638. , nan, nan, nan, nan, nan, nan, nan], [1638. , nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - intensity_rtnrt(time, site)float640.0 nan nan nan ... nan nan nan nan
- long_name :
- Rain intensity based upon accum_rtnrt
- units :
- mm/hr
- _FillValue :
- -999999.0
- valid_min :
- 0.0
- valid_max :
- 30000.0
- threshold :
- 0.3 mm/hr
- absolute_accuracy :
- plus/minus 6
- equation :
- Calculated by accum_rtnrt * 60
- comment :
- Only measurements that exceed the threshold are recorded. Any measurement below the threshold is reported as 0 mm/hr.
- source :
- gucwbpluvio2M1.a1
array([[ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 7.72800011, nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [12.67199982, nan, nan, nan, nan, nan, nan, nan], [ 3.7119999 , nan, nan, nan, nan, nan, nan, nan], ... [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - atmos_pressure(time, site)float64354.9 nan nan nan ... nan nan nan
- long_name :
- Atmospheric pressure
- units :
- kPa
- _FillValue :
- -999999.0
- standard_name :
- surface_air_pressure
- source :
- gucmetM1.b1
array([[354.91715271, nan, nan, nan, nan, nan, nan, nan], [354.97465739, nan, nan, nan, nan, nan, nan, nan], [355.05001831, nan, nan, nan, nan, nan, nan, nan], [355. , nan, nan, nan, nan, nan, nan, nan], [355. , nan, nan, nan, nan, nan, nan, nan], [355.01548177, nan, nan, nan, nan, nan, nan, nan], [355. , nan, nan, nan, nan, nan, nan, nan], [355. , nan, nan, nan, nan, nan, nan, nan], [355.08836792, nan, nan, nan, nan, nan, nan, nan], [355.09997559, nan, nan, nan, nan, nan, nan, nan], ... [359.8500061 , nan, nan, nan, nan, nan, nan, nan], [359.84249878, nan, nan, nan, nan, nan, nan, nan], [359.81364695, nan, nan, nan, nan, nan, nan, nan], [359.78997803, nan, nan, nan, nan, nan, nan, nan], [359.83333588, nan, nan, nan, nan, nan, nan, nan], [359.88585256, nan, nan, nan, nan, nan, nan, nan], [359.93548126, nan, nan, nan, nan, nan, nan, nan], [359.98582204, nan, nan, nan, nan, nan, nan, nan], [360.03322591, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - temp_mean(time, site)float643.09 nan nan nan ... nan nan nan
- long_name :
- Temperature mean
- units :
- degC
- _FillValue :
- -999999.0
- standard_name :
- air_temperature
- source :
- gucmetM1.b1
array([[ 3.09038353, nan, nan, nan, nan, nan, nan, nan], [ -1.54097332, nan, nan, nan, nan, nan, nan, nan], [ -6.43320009, nan, nan, nan, nan, nan, nan, nan], [ -9.07520023, nan, nan, nan, nan, nan, nan, nan], [ -9.05183994, nan, nan, nan, nan, nan, nan, nan], [ -9.0999199 , nan, nan, nan, nan, nan, nan, nan], [-10.46672035, nan, nan, nan, nan, nan, nan, nan], [-10.85595947, nan, nan, nan, nan, nan, nan, nan], [ -9.90187996, nan, nan, nan, nan, nan, nan, nan], [ -9.79073263, nan, nan, nan, nan, nan, nan, nan], ... [ 12.79389942, nan, nan, nan, nan, nan, nan, nan], [ 11.44999981, nan, nan, nan, nan, nan, nan, nan], [ 12.63298343, nan, nan, nan, nan, nan, nan, nan], [ 13.31370072, nan, nan, nan, nan, nan, nan, nan], [ 11.3607498 , nan, nan, nan, nan, nan, nan, nan], [ 6.90031637, nan, nan, nan, nan, nan, nan, nan], [ 3.06794999, nan, nan, nan, nan, nan, nan, nan], [ 0.63583322, nan, nan, nan, nan, nan, nan, nan], [ 0.31454667, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - temp_std(time, site)float640.2666 nan nan nan ... nan nan nan
- long_name :
- Temperature standard deviation
- units :
- degC
- _FillValue :
- -999999.0
- source :
- gucmetM1.b1
array([[0.26657334, nan, nan, nan, nan, nan, nan, nan], [0.26691999, nan, nan, nan, nan, nan, nan, nan], [0.20532667, nan, nan, nan, nan, nan, nan, nan], [0.22641332, nan, nan, nan, nan, nan, nan, nan], [0.21723999, nan, nan, nan, nan, nan, nan, nan], [0.21973332, nan, nan, nan, nan, nan, nan, nan], [0.21752001, nan, nan, nan, nan, nan, nan, nan], [0.21688001, nan, nan, nan, nan, nan, nan, nan], [0.22648 , nan, nan, nan, nan, nan, nan, nan], [0.21313333, nan, nan, nan, nan, nan, nan, nan], ... [0.25510001, nan, nan, nan, nan, nan, nan, nan], [0.32824999, nan, nan, nan, nan, nan, nan, nan], [0.27086667, nan, nan, nan, nan, nan, nan, nan], [0.22709999, nan, nan, nan, nan, nan, nan, nan], [0.24075001, nan, nan, nan, nan, nan, nan, nan], [0.29553333, nan, nan, nan, nan, nan, nan, nan], [0.3009 , nan, nan, nan, nan, nan, nan, nan], [0.3380833 , nan, nan, nan, nan, nan, nan, nan], [0.25832002, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - rh_mean(time, site)float64308.4 nan nan nan ... nan nan nan
- long_name :
- Relative humidity mean
- units :
- %
- _FillValue :
- -999999.0
- standard_name :
- relative_humidity
- source :
- gucmetM1.b1
array([[308.40584798, nan, nan, nan, nan, nan, nan, nan], [353.1053536 , nan, nan, nan, nan, nan, nan, nan], [426.35002309, nan, nan, nan, nan, nan, nan, nan], [458.05599325, nan, nan, nan, nan, nan, nan, nan], [459.59197632, nan, nan, nan, nan, nan, nan, nan], [461.70932943, nan, nan, nan, nan, nan, nan, nan], [477.59999878, nan, nan, nan, nan, nan, nan, nan], [485.84396606, nan, nan, nan, nan, nan, nan, nan], [479.9319873 , nan, nan, nan, nan, nan, nan, nan], [478.64798828, nan, nan, nan, nan, nan, nan, nan], ... [127.6805069 , nan, nan, nan, nan, nan, nan, nan], [149.06499481, nan, nan, nan, nan, nan, nan, nan], [146.13866018, nan, nan, nan, nan, nan, nan, nan], [145.74949951, nan, nan, nan, nan, nan, nan, nan], [167.57334518, nan, nan, nan, nan, nan, nan, nan], [193.37132924, nan, nan, nan, nan, nan, nan, nan], [202.91649017, nan, nan, nan, nan, nan, nan, nan], [213.0858256 , nan, nan, nan, nan, nan, nan, nan], [220.91518311, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - rh_std(time, site)float642.404 nan nan nan ... nan nan nan
- long_name :
- Relative humidity standard deviation
- units :
- %
- _FillValue :
- -999999.0
- source :
- gucmetM1.b1
array([[2.40418996, nan, nan, nan, nan, nan, nan, nan], [2.28075986, nan, nan, nan, nan, nan, nan, nan], [1.17251332, nan, nan, nan, nan, nan, nan, nan], [0.71521332, nan, nan, nan, nan, nan, nan, nan], [0.58603999, nan, nan, nan, nan, nan, nan, nan], [0.60641332, nan, nan, nan, nan, nan, nan, nan], [0.65215997, nan, nan, nan, nan, nan, nan, nan], [0.59043996, nan, nan, nan, nan, nan, nan, nan], [0.61148 , nan, nan, nan, nan, nan, nan, nan], [0.66414666, nan, nan, nan, nan, nan, nan, nan], ... [1.47390003, nan, nan, nan, nan, nan, nan, nan], [1.97849989, nan, nan, nan, nan, nan, nan, nan], [1.26928333, nan, nan, nan, nan, nan, nan, nan], [0.96059997, nan, nan, nan, nan, nan, nan, nan], [0.80499999, nan, nan, nan, nan, nan, nan, nan], [1.00775007, nan, nan, nan, nan, nan, nan, nan], [0.71995 , nan, nan, nan, nan, nan, nan, nan], [0.79683335, nan, nan, nan, nan, nan, nan, nan], [0.58834663, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - vapor_pressure_mean(time, site)float641.97 nan nan nan ... nan nan nan
- long_name :
- Vapor pressure mean, calculated
- units :
- kPa
- _FillValue :
- -999999.0
- standard_name :
- water_vapor_partial_pressure_in_air
- source :
- gucmetM1.b1
array([[1.96957995, nan, nan, nan, nan, nan, nan, nan], [2.09102667, nan, nan, nan, nan, nan, nan, nan], [2.32710325, nan, nan, nan, nan, nan, nan, nan], [2.39158664, nan, nan, nan, nan, nan, nan, nan], [2.39979986, nan, nan, nan, nan, nan, nan, nan], [2.41018658, nan, nan, nan, nan, nan, nan, nan], [2.43544002, nan, nan, nan, nan, nan, nan, nan], [2.46152011, nan, nan, nan, nan, nan, nan, nan], [2.46992002, nan, nan, nan, nan, nan, nan, nan], [2.46930669, nan, nan, nan, nan, nan, nan, nan], ... [0.93798333, nan, nan, nan, nan, nan, nan, nan], [1.07350001, nan, nan, nan, nan, nan, nan, nan], [1.07053333, nan, nan, nan, nan, nan, nan, nan], [1.07805003, nan, nan, nan, nan, nan, nan, nan], [1.20441666, nan, nan, nan, nan, nan, nan, nan], [1.30396658, nan, nan, nan, nan, nan, nan, nan], [1.29534992, nan, nan, nan, nan, nan, nan, nan], [1.31141663, nan, nan, nan, nan, nan, nan, nan], [1.35394675, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - vapor_pressure_std(time, site)float640.01522 nan nan nan ... nan nan nan
- long_name :
- Vapor pressure standard deviation
- units :
- kPa
- _FillValue :
- -999999.0
- source :
- gucmetM1.b1
array([[0.01521667, nan, nan, nan, nan, nan, nan, nan], [0.01150667, nan, nan, nan, nan, nan, nan, nan], [0.01075667, nan, nan, nan, nan, nan, nan, nan], [0.01 , nan, nan, nan, nan, nan, nan, nan], [0.01 , nan, nan, nan, nan, nan, nan, nan], [0.01 , nan, nan, nan, nan, nan, nan, nan], [0.01 , nan, nan, nan, nan, nan, nan, nan], [0.01 , nan, nan, nan, nan, nan, nan, nan], [0.01 , nan, nan, nan, nan, nan, nan, nan], [0.01 , nan, nan, nan, nan, nan, nan, nan], ... [0.01146667, nan, nan, nan, nan, nan, nan, nan], [0.0135 , nan, nan, nan, nan, nan, nan, nan], [0.00868333, nan, nan, nan, nan, nan, nan, nan], [0.00745 , nan, nan, nan, nan, nan, nan, nan], [0.00541667, nan, nan, nan, nan, nan, nan, nan], [0.006 , nan, nan, nan, nan, nan, nan, nan], [0.005 , nan, nan, nan, nan, nan, nan, nan], [0.00775 , nan, nan, nan, nan, nan, nan, nan], [0.006 , nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - wspd_arith_mean(time, site)float6412.35 nan nan nan ... nan nan nan
- long_name :
- Wind speed arithmetic mean
- units :
- m/s
- _FillValue :
- -999999.0
- source :
- gucmetM1.b1
array([[12.34554363, nan, nan, nan, nan, nan, nan, nan], [11.18824053, nan, nan, nan, nan, nan, nan, nan], [ 2.80318997, nan, nan, nan, nan, nan, nan, nan], [ 7.82461306, nan, nan, nan, nan, nan, nan, nan], [ 4.53800005, nan, nan, nan, nan, nan, nan, nan], [ 4.53325343, nan, nan, nan, nan, nan, nan, nan], [ 4.01096006, nan, nan, nan, nan, nan, nan, nan], [ 4.41987986, nan, nan, nan, nan, nan, nan, nan], [ 6.41915983, nan, nan, nan, nan, nan, nan, nan], [ 5.94351977, nan, nan, nan, nan, nan, nan, nan], ... [10.91038362, nan, nan, nan, nan, nan, nan, nan], [14.81475019, nan, nan, nan, nan, nan, nan, nan], [13.05993341, nan, nan, nan, nan, nan, nan, nan], [16.66340017, nan, nan, nan, nan, nan, nan, nan], [18.71866814, nan, nan, nan, nan, nan, nan, nan], [17.24324961, nan, nan, nan, nan, nan, nan, nan], [11.30305018, nan, nan, nan, nan, nan, nan, nan], [10.06558299, nan, nan, nan, nan, nan, nan, nan], [13.76426699, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - wspd_vec_mean(time, site)float6411.9 nan nan nan ... nan nan nan
- long_name :
- Wind speed vector mean
- units :
- m/s
- _FillValue :
- -999999.0
- source :
- gucmetM1.b1
array([[11.90151355, nan, nan, nan, nan, nan, nan, nan], [11.03848016, nan, nan, nan, nan, nan, nan, nan], [ 2.59260342, nan, nan, nan, nan, nan, nan, nan], [ 7.80234664, nan, nan, nan, nan, nan, nan, nan], [ 4.50368021, nan, nan, nan, nan, nan, nan, nan], [ 4.51602653, nan, nan, nan, nan, nan, nan, nan], [ 3.95744 , nan, nan, nan, nan, nan, nan, nan], [ 4.36195995, nan, nan, nan, nan, nan, nan, nan], [ 6.27535938, nan, nan, nan, nan, nan, nan, nan], [ 5.67589312, nan, nan, nan, nan, nan, nan, nan], ... [10.74444931, nan, nan, nan, nan, nan, nan, nan], [14.72975063, nan, nan, nan, nan, nan, nan, nan], [12.94308292, nan, nan, nan, nan, nan, nan, nan], [16.53030024, nan, nan, nan, nan, nan, nan, nan], [18.55991729, nan, nan, nan, nan, nan, nan, nan], [17.07980083, nan, nan, nan, nan, nan, nan, nan], [11.19969959, nan, nan, nan, nan, nan, nan, nan], [ 9.95574991, nan, nan, nan, nan, nan, nan, nan], [13.4548002 , nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - wdir_vec_mean(time, site)float641.015e+03 nan nan ... nan nan nan
- long_name :
- Wind direction vector mean
- units :
- degree
- _FillValue :
- -999999.0
- standard_name :
- wind_from_direction
- source :
- gucmetM1.b1
array([[1014.53865967, nan, nan, nan, nan, nan, nan, nan], [1148.81733398, nan, nan, nan, nan, nan, nan, nan], [ 706.89902588, nan, nan, nan, nan, nan, nan, nan], [1538.63200521, nan, nan, nan, nan, nan, nan, nan], [1528.32804687, nan, nan, nan, nan, nan, nan, nan], [ 460.20413167, nan, nan, nan, nan, nan, nan, nan], [1160.92 , nan, nan, nan, nan, nan, nan, nan], [1230.55201172, nan, nan, nan, nan, nan, nan, nan], [1662.87835449, nan, nan, nan, nan, nan, nan, nan], [ 498.11467855, nan, nan, nan, nan, nan, nan, nan], ... [1438.13671265, nan, nan, nan, nan, nan, nan, nan], [1352.82504272, nan, nan, nan, nan, nan, nan, nan], [1529.61840007, nan, nan, nan, nan, nan, nan, nan], [1639.91997681, nan, nan, nan, nan, nan, nan, nan], [1640.74996948, nan, nan, nan, nan, nan, nan, nan], [1620.87173665, nan, nan, nan, nan, nan, nan, nan], [1603.91998901, nan, nan, nan, nan, nan, nan, nan], [1591.39162191, nan, nan, nan, nan, nan, nan, nan], [1526.34404622, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - wdir_vec_std(time, site)float6471.69 nan nan nan ... nan nan nan
- long_name :
- Wind direction vector mean standard deviation
- units :
- degree
- _FillValue :
- -999999.0
- source :
- gucmetM1.b1
array([[ 71.68630033, nan, nan, nan, nan, nan, nan, nan], [ 43.30753286, nan, nan, nan, nan, nan, nan, nan], [140.61893723, nan, nan, nan, nan, nan, nan, nan], [ 21.04652033, nan, nan, nan, nan, nan, nan, nan], [ 34.21868179, nan, nan, nan, nan, nan, nan, nan], [ 26.1570787 , nan, nan, nan, nan, nan, nan, nan], [ 39.69831757, nan, nan, nan, nan, nan, nan, nan], [ 41.87564152, nan, nan, nan, nan, nan, nan, nan], [ 67.79447632, nan, nan, nan, nan, nan, nan, nan], [ 83.92852743, nan, nan, nan, nan, nan, nan, nan], ... [ 48.08993047, nan, nan, nan, nan, nan, nan, nan], [ 29.54649973, nan, nan, nan, nan, nan, nan, nan], [ 38.03831746, nan, nan, nan, nan, nan, nan, nan], [ 34.59799938, nan, nan, nan, nan, nan, nan, nan], [ 37.40250174, nan, nan, nan, nan, nan, nan, nan], [ 38.44693222, nan, nan, nan, nan, nan, nan, nan], [ 37.49799995, nan, nan, nan, nan, nan, nan, nan], [ 40.72358513, nan, nan, nan, nan, nan, nan, nan], [ 57.64311025, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - pwd_err_code(time, site)float640.0 nan nan nan ... nan nan nan nan
- long_name :
- PWD alarm
- units :
- 1
- _FillValue :
- -999999.0
- source :
- gucmetM1.b1
array([[ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], ... [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan]]) - pwd_mean_vis_1min(time, site)float647.201e+04 nan nan ... nan nan nan
- long_name :
- PWD 1 minute mean visibility
- units :
- m
- _FillValue :
- -999999.0
- standard_name :
- visibility_in_air
- source :
- gucmetM1.b1
array([[ 72013.8 , nan, nan, nan, nan, nan, nan, nan], [ 14220.28 , nan, nan, nan, nan, nan, nan, nan], [ 10177.28 , nan, nan, nan, nan, nan, nan, nan], [ 15722.42666667, nan, nan, nan, nan, nan, nan, nan], [ 22775.12 , nan, nan, nan, nan, nan, nan, nan], [ 7851.28 , nan, nan, nan, nan, nan, nan, nan], [ 5536.8 , nan, nan, nan, nan, nan, ... nan, nan, nan, nan, nan], [ 99387.08333333, nan, nan, nan, nan, nan, nan, nan], [100000. , nan, nan, nan, nan, nan, nan, nan], [100000. , nan, nan, nan, nan, nan, nan, nan], [100000. , nan, nan, nan, nan, nan, nan, nan], [100000. , nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - pwd_mean_vis_10min(time, site)float649.271e+04 nan nan ... nan nan nan
- long_name :
- PWD 10 minute mean visibility
- units :
- m
- _FillValue :
- -999999.0
- standard_name :
- visibility_in_air
- source :
- gucmetM1.b1
array([[ 92709.37666667, nan, nan, nan, nan, nan, nan, nan], [ 17871.26666667, nan, nan, nan, nan, nan, nan, nan], [ 11428.81666667, nan, nan, nan, nan, nan, nan, nan], [ 8147.28 , nan, nan, nan, nan, nan, nan, nan], [ 14005.2 , nan, nan, nan, nan, nan, nan, nan], [ 11877.02666667, nan, nan, nan, nan, nan, nan, nan], [ 4909.56 , nan, nan, nan, nan, nan, ... nan, nan, nan, nan, nan], [100000. , nan, nan, nan, nan, nan, nan, nan], [100000. , nan, nan, nan, nan, nan, nan, nan], [100000. , nan, nan, nan, nan, nan, nan, nan], [100000. , nan, nan, nan, nan, nan, nan, nan], [100000. , nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - pwd_pw_code_inst(time, site)float64162.3 nan nan nan ... nan nan nan
- long_name :
- PWD instantaneous present weather code
- units :
- 1
- _FillValue :
- -999999.0
- source :
- gucmetM1.b1
array([[162.29333333, nan, nan, nan, nan, nan, nan, nan], [355. , nan, nan, nan, nan, nan, nan, nan], [355. , nan, nan, nan, nan, nan, nan, nan], [355. , nan, nan, nan, nan, nan, nan, nan], [355. , nan, nan, nan, nan, nan, nan, nan], [355. , nan, nan, nan, nan, nan, nan, nan], [358.88 , nan, nan, nan, nan, nan, nan, nan], [355. , nan, nan, nan, nan, nan, nan, nan], [358.84 , nan, nan, nan, nan, nan, nan, nan], [360.02666667, nan, nan, nan, nan, nan, nan, nan], ... [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - pwd_pw_code_15min(time, site)float64251.5 nan nan nan ... nan nan nan
- long_name :
- PWD 15 minute present weather code
- units :
- 1
- _FillValue :
- -999999.0
- source :
- gucmetM1.b1
array([[251.52333333, nan, nan, nan, nan, nan, nan, nan], [425. , nan, nan, nan, nan, nan, nan, nan], [425. , nan, nan, nan, nan, nan, nan, nan], [425. , nan, nan, nan, nan, nan, nan, nan], [425. , nan, nan, nan, nan, nan, nan, nan], [425. , nan, nan, nan, nan, nan, nan, nan], [430. , nan, nan, nan, nan, nan, nan, nan], [426.2 , nan, nan, nan, nan, nan, nan, nan], [430. , nan, nan, nan, nan, nan, nan, nan], [430. , nan, nan, nan, nan, nan, nan, nan], ... [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - pwd_pw_code_1hr(time, site)float64144.5 nan nan nan ... nan nan nan
- long_name :
- PWD 1 hour present weather code
- units :
- 1
- _FillValue :
- -999999.0
- source :
- gucmetM1.b1
array([[144.45 , nan, nan, nan, nan, nan, nan, nan], [425. , nan, nan, nan, nan, nan, nan, nan], [425. , nan, nan, nan, nan, nan, nan, nan], [425. , nan, nan, nan, nan, nan, nan, nan], [425. , nan, nan, nan, nan, nan, nan, nan], [425. , nan, nan, nan, nan, nan, nan, nan], [430. , nan, nan, nan, nan, nan, nan, nan], [430. , nan, nan, nan, nan, nan, nan, nan], [430. , nan, nan, nan, nan, nan, nan, nan], [430. , nan, nan, nan, nan, nan, nan, nan], ... [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - pwd_precip_rate_mean_1min(time, site)float641.287 nan nan nan ... nan nan nan
- long_name :
- PWD 1 minute mean precipitation rate
- units :
- mm/hr
- _FillValue :
- -999999.0
- standard_name :
- lwe_precipitation_rate
- source :
- gucmetM1.b1
array([[1.28706661e+00, nan, nan, nan, nan, nan, nan, nan], [4.05813339e+00, nan, nan, nan, nan, nan, nan, nan], [1.93503334e+00, nan, nan, nan, nan, nan, nan, nan], [2.43413310e+00, nan, nan, nan, nan, nan, nan, nan], [3.68000054e-01, nan, nan, nan, nan, nan, nan, nan], [4.35733324e+00, nan, nan, nan, nan, nan, nan, nan], [5.87760029e+00, nan, nan, nan, nan, nan, nan, nan], [2.41159997e+00, nan, nan, nan, nan, nan, nan, nan], [3.00080000e+00, nan, nan, nan, nan, nan, nan, nan], [9.65119947e+00, nan, nan, nan, nan, nan, nan, nan], ... [0.00000000e+00, nan, nan, nan, nan, nan, nan, nan], [0.00000000e+00, nan, nan, nan, nan, nan, nan, nan], [0.00000000e+00, nan, nan, nan, nan, nan, nan, nan], [0.00000000e+00, nan, nan, nan, nan, nan, nan, nan], [0.00000000e+00, nan, nan, nan, nan, nan, nan, nan], [0.00000000e+00, nan, nan, nan, nan, nan, nan, nan], [0.00000000e+00, nan, nan, nan, nan, nan, nan, nan], [0.00000000e+00, nan, nan, nan, nan, nan, nan, nan], [0.00000000e+00, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - pwd_cumul_rain(time, site)float64418.5 nan nan nan ... nan nan nan
- long_name :
- PWD cumulative liquid precipitation
- units :
- mm
- _FillValue :
- -999999.0
- source :
- gucmetM1.b1
array([[418.47854075, nan, nan, nan, nan, nan, nan, nan], [419.04801839, nan, nan, nan, nan, nan, nan, nan], [419.92595174, nan, nan, nan, nan, nan, nan, nan], [420.78053792, nan, nan, nan, nan, nan, nan, nan], [420.89679443, nan, nan, nan, nan, nan, nan, nan], [421.29971558, nan, nan, nan, nan, nan, nan, nan], [424.33319092, nan, nan, nan, nan, nan, nan, nan], [425.16201172, nan, nan, nan, nan, nan, nan, nan], [426.07078735, nan, nan, nan, nan, nan, nan, nan], [426.67426066, nan, nan, nan, nan, nan, nan, nan], ... [462.29998779, nan, nan, nan, nan, nan, nan, nan], [462.29998779, nan, nan, nan, nan, nan, nan, nan], [462.29998779, nan, nan, nan, nan, nan, nan, nan], [462.29998779, nan, nan, nan, nan, nan, nan, nan], [462.29998779, nan, nan, nan, nan, nan, nan, nan], [462.29998779, nan, nan, nan, nan, nan, nan, nan], [462.29998779, nan, nan, nan, nan, nan, nan, nan], [462.29998779, nan, nan, nan, nan, nan, nan, nan], [462.29998779, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - pwd_cumul_snow(time, site)float644.176e+03 nan nan ... nan nan nan
- long_name :
- PWD cumulative snow
- units :
- mm
- _FillValue :
- -999999.0
- source :
- gucmetM1.b1
array([[4175.71333333, nan, nan, nan, nan, nan, nan, nan], [4181.97333333, nan, nan, nan, nan, nan, nan, nan], [4190.75666667, nan, nan, nan, nan, nan, nan, nan], [4199.68 , nan, nan, nan, nan, nan, nan, nan], [4200. , nan, nan, nan, nan, nan, nan, nan], [4205.90666667, nan, nan, nan, nan, nan, nan, nan], [4235.8 , nan, nan, nan, nan, nan, nan, nan], [4244.28 , nan, nan, nan, nan, nan, nan, nan], [4252.88 , nan, nan, nan, nan, nan, nan, nan], [4258.29333333, nan, nan, nan, nan, nan, nan, nan], ... [4615. , nan, nan, nan, nan, nan, nan, nan], [4615. , nan, nan, nan, nan, nan, nan, nan], [4615. , nan, nan, nan, nan, nan, nan, nan], [4615. , nan, nan, nan, nan, nan, nan, nan], [4615. , nan, nan, nan, nan, nan, nan, nan], [4615. , nan, nan, nan, nan, nan, nan, nan], [4615. , nan, nan, nan, nan, nan, nan, nan], [4615. , nan, nan, nan, nan, nan, nan, nan], [4615. , nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - org_precip_rate_mean(time, site)float640.1068 nan nan nan ... nan nan nan
- long_name :
- ORG precipitation rate mean
- units :
- mm/hr
- _FillValue :
- -999999.0
- standard_name :
- lwe_precipitation_rate
- source :
- gucmetM1.b1
array([[1.06803327e-01, nan, nan, nan, nan, nan, nan, nan], [9.53653369e-01, nan, nan, nan, nan, nan, nan, nan], [7.84413321e-01, nan, nan, nan, nan, nan, nan, nan], [3.67213345e-01, nan, nan, nan, nan, nan, nan, nan], [9.11599922e-02, nan, nan, nan, nan, nan, nan, nan], [1.91324006e+00, nan, nan, nan, nan, nan, nan, nan], [1.72952000e+00, nan, nan, nan, nan, nan, nan, nan], [7.55559969e-01, nan, nan, nan, nan, nan, nan, nan], [1.21340000e+00, nan, nan, nan, nan, nan, nan, nan], [4.55558675e+00, nan, nan, nan, nan, nan, nan, nan], ... [0.00000000e+00, nan, nan, nan, nan, nan, nan, nan], [0.00000000e+00, nan, nan, nan, nan, nan, nan, nan], [0.00000000e+00, nan, nan, nan, nan, nan, nan, nan], [0.00000000e+00, nan, nan, nan, nan, nan, nan, nan], [0.00000000e+00, nan, nan, nan, nan, nan, nan, nan], [0.00000000e+00, nan, nan, nan, nan, nan, nan, nan], [0.00000000e+00, nan, nan, nan, nan, nan, nan, nan], [0.00000000e+00, nan, nan, nan, nan, nan, nan, nan], [0.00000000e+00, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - tbrg_precip_total(time, site)float640.0 nan nan nan ... nan nan nan nan
- long_name :
- TBRG precipitation total
- units :
- mm
- _FillValue :
- -999999.0
- source :
- gucmetM1.b1
array([[0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], ... [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [0. , nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - tbrg_precip_total_corr(time, site)float640.0 nan nan nan ... nan nan nan nan
- long_name :
- TBRG precipitation total, corrected
- units :
- mm
- _FillValue :
- -999999.0
- source :
- gucmetM1.b1
array([[ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], ... [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ 0. , nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - precip_rate(time, site)float640.6511 0.0 nan nan ... nan nan nan
- long_name :
- Precipitation intensity
- units :
- mm/hr
- _FillValue :
- -999999.0
- standard_name :
- lwe_precipitation_rate
- source :
- gucldM1.b1
array([[6.51086689e-01, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [1.00385333e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [1.80504661e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [1.69600004e-01, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [1.59900000e+00, 7.72440046e-01, nan, nan, nan, nan, nan, nan], [3.06888006e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [2.50075987e+00, 3.55047981e+00, nan, nan, nan, nan, nan, nan], [5.62396017e+00, 1.50910399e+01, nan, nan, nan, nan, nan, nan], [7.90029304e+00, 8.64768002e+00, nan, nan, nan, nan, nan, nan], ... [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - weather_code(time, site)float64172.3 0.0 nan nan ... nan nan nan
- long_name :
- SYNOP WaWa Table 4680
- units :
- 1
- _FillValue :
- -999999.0
- source :
- gucldM1.b1
array([[172.29333333, 0. , nan, nan, nan, nan, nan, nan], [247.08 , 0. , nan, nan, nan, nan, nan, nan], [355. , 0. , nan, nan, nan, nan, nan, nan], [ 30.29333333, 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [355. , 135.37333333, nan, nan, nan, nan, nan, nan], [355. , 0. , nan, nan, nan, nan, nan, nan], [355. , 252.2 , nan, nan, nan, nan, nan, nan], [359.84 , 360.8 , nan, nan, nan, nan, nan, nan], [360. , 360. , nan, nan, nan, nan, nan, nan], ... [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - number_detected_particles(time, site)float64434.1 0.0 nan nan ... nan nan nan
- long_name :
- Number of particles detected
- units :
- count
- _FillValue :
- -999999.0
- source :
- gucldM1.b1
array([[4.34066667e+02, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [8.56893333e+02, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [1.38405000e+03, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [8.00000000e+01, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [1.84457333e+03, 5.85253333e+02, nan, nan, nan, nan, nan, nan], [2.64280000e+03, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [2.35940000e+03, 2.02976000e+03, nan, nan, nan, nan, nan, nan], [4.23356000e+03, 6.81076000e+03, nan, nan, nan, nan, nan, nan], [5.86906667e+03, 5.18493333e+03, nan, nan, nan, nan, nan, nan], ... [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - mor_visibility(time, site)float644.37e+04 9.553e+04 nan ... nan nan
- long_name :
- Meteorological optical range visibility
- units :
- m
- _FillValue :
- -999999.0
- standard_name :
- visibility_in_air
- source :
- gucldM1.b1
array([[ 43703.63333333, 95532.59333333, nan, nan, nan, nan, nan, nan], [ 8397.68 , 71145.4 , nan, nan, nan, nan, nan, nan], [ 4612.71 , 99999.75666667, nan, nan, nan, nan, nan, nan], [ 11361.32 , 100000. , nan, nan, nan, nan, nan, nan], [ 21220.52 , 100000. , nan, nan, nan, nan, nan, nan], [ 3567.76 , 29193.70666667, nan, nan, nan, nan, nan, nan], [ 3096.32 , 47570.48 , nan, nan, nan, nan, ... nan, nan, nan, nan, nan], [100000. , 100000. , nan, nan, nan, nan, nan, nan], [100000. , 100000. , nan, nan, nan, nan, nan, nan], [100000. , 100000. , nan, nan, nan, nan, nan, nan], [100000. , 100000. , nan, nan, nan, nan, nan, nan], [100000. , 100000. , nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - snow_depth_intensity(time, site)float644.14 0.0 nan nan ... nan nan nan
- long_name :
- New snow height
- units :
- mm/hr
- _FillValue :
- -999999.0
- comment :
- This value is valid on a short period of one hour and its purpose is to provide new snow height on railways or roads for the purposes of safety. It is not equivalent to the WMO definition of snow intensity nor does if follow from WMO observation guide lines.
- source :
- gucldM1.b1
array([[ 4.14 , 0. , nan, nan, nan, nan, nan, nan], [ 6.96 , 0. , nan, nan, nan, nan, nan, nan], [ 41.80666667, 0. , nan, nan, nan, nan, nan, nan], [ 5.12 , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 26.89333333, 1.68 , nan, nan, nan, nan, nan, nan], [ 25.6 , 0. , nan, nan, nan, nan, nan, nan], [ 18.84 , 7.08 , nan, nan, nan, nan, nan, nan], [ 26.4 , 24.52 , nan, nan, nan, nan, nan, nan], [ 44.17333333, 15.24 , nan, nan, nan, nan, nan, nan], ... [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - class_size_width(time, site, particle_size)float640.125 0.125 0.125 ... nan nan nan
- long_name :
- Class size width
- units :
- mm
- _FillValue :
- -999999.0
- source :
- gucldM1.b1
array([[[0.125, 0.125, 0.125, ..., 2. , 3. , 3. ], [0.125, 0.125, 0.125, ..., 2. , 3. , 3. ], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[0.125, 0.125, 0.125, ..., 2. , 3. , 3. ], [0.125, 0.125, 0.125, ..., 2. , 3. , 3. ], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[0.125, 0.125, 0.125, ..., 2. , 3. , 3. ], [0.125, 0.125, 0.125, ..., 2. , 3. , 3. ], [ nan, nan, nan, ..., nan, nan, nan], ..., ... ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[0.125, 0.125, 0.125, ..., 2. , 3. , 3. ], [0.125, 0.125, 0.125, ..., 2. , 3. , 3. ], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - fall_velocity_calculated(time, site, raw_fall_velocity)float640.277 0.822 1.351 ... nan nan nan
- long_name :
- Fall velocity calculated after Lhermite
- units :
- m/s
- _FillValue :
- -999999.0
- source :
- gucldM1.b1
array([[[0.27700001, 0.82200003, 1.35099995, ..., 9.19999981, 9.19999981, 9.19999981], [0.27700001, 0.82200003, 1.35099995, ..., 9.19999981, 9.19999981, 9.19999981], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[0.27700001, 0.82200003, 1.35099995, ..., 9.19999981, 9.19999981, 9.19999981], [0.27700001, 0.82200003, 1.35099995, ..., 9.19999981, 9.19999981, 9.19999981], [ nan, nan, nan, ..., nan, nan, nan], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - raw_spectrum(time, site, particle_size, raw_fall_velocity)float640.0 0.0 0.0 0.0 ... nan nan nan nan
- long_name :
- Raw drop size distribution
- units :
- count
- _FillValue :
- -999999.0
- source :
- gucldM1.b1
array([[[[ 0., 0., 0., ..., 0., 0., 0.], [ 0., 0., 0., ..., 0., 0., 0.], [ 0., 0., 0., ..., 0., 0., 0.], ..., [ 0., 0., 0., ..., 0., 0., 0.], [ 0., 0., 0., ..., 0., 0., 0.], [ 0., 0., 0., ..., 0., 0., 0.]], [[ 0., 0., 0., ..., 0., 0., 0.], [ 0., 0., 0., ..., 0., 0., 0.], [ 0., 0., 0., ..., 0., 0., 0.], ..., [ 0., 0., 0., ..., 0., 0., 0.], [ 0., 0., 0., ..., 0., 0., 0.], [ 0., 0., 0., ..., 0., 0., 0.]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., ... ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]]]) - liquid_water_content(time, site)float64312.3 0.0 nan nan ... nan nan nan
- long_name :
- Liquid water content
- units :
- mm^3/m^3
- _FillValue :
- -999999.0
- source :
- gucldM1.b1
array([[3.12348969e+02, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [6.32090449e+02, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [2.62446473e+03, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [3.06777109e+02, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [1.96168814e+03, 2.07699032e+02, nan, nan, nan, nan, nan, nan], [2.22793631e+03, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [1.63764633e+03, 7.59445455e+02, nan, nan, nan, nan, nan, nan], [2.44384792e+03, 2.68750806e+03, nan, nan, nan, nan, nan, nan], [3.95109573e+03, 1.82020264e+03, nan, nan, nan, nan, nan, nan], ... [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - equivalent_radar_reflectivity(time, site)float6497.04 0.0 nan nan ... nan nan nan
- long_name :
- Radar reflectivity calculated by the ingest
- units :
- dBZ
- _FillValue :
- -999999.0
- source :
- gucldM1.b1
array([[ 9.70357459e+01, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [ 1.33962568e+02, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [ 2.31198292e+02, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [ 2.26871859e+01, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [ 0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [ 2.23034760e+02, 5.65289728e+01, nan, nan, nan, nan, nan, nan], [ 2.08531131e+02, 0.00000000e+00, nan, nan, nan, nan, ... nan, nan, nan, nan, nan], [ 0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [ 0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [ 0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [ 0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [ 0.00000000e+00, 0.00000000e+00, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - intercept_parameter(time, site)float64623.9 0.0 nan nan ... nan nan nan
- long_name :
- Intercept parameter, assuming an ideal Marshall-Palmer type distribution
- units :
- 1/(m^3 mm)
- _FillValue :
- -999999.0
- source :
- gucldM1.b1
array([[ 623.91405629, 0. , nan, nan, nan, nan, nan, nan], [ 2599.37180501, 0. , nan, nan, nan, nan, nan, nan], [ 5219.2559375 , 0. , nan, nan, nan, nan, nan, nan], [ 48.61558268, 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 3443.29441732, 6217.69508464, nan, nan, nan, nan, nan, nan], [ 9475.95332031, 0. , nan, nan, nan, nan, nan, nan], [ 7242.7003125 , 11106.74410156, nan, nan, nan, nan, nan, nan], [27708.265625 , 42622.610625 , nan, nan, nan, nan, nan, nan], [20520.53497396, 38593.33619792, nan, nan, nan, nan, nan, nan], ... [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - slope_parameter(time, site)float643.596 0.0 nan nan ... nan nan nan
- long_name :
- Slope parameter, assuming an ideal Marshall-Palmer type distribution
- units :
- 1/mm
- _FillValue :
- -999999.0
- source :
- gucldM1.b1
array([[ 3.59618416, 0. , nan, nan, nan, nan, nan, nan], [ 6.37153338, 0. , nan, nan, nan, nan, nan, nan], [ 7.68033747, 0. , nan, nan, nan, nan, nan, nan], [ 0.37863337, 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 7.42953428, 5.78833961, nan, nan, nan, nan, nan, nan], [ 9.51272865, 0. , nan, nan, nan, nan, nan, nan], [ 9.71870224, 9.35737396, nan, nan, nan, nan, nan, nan], [11.52822281, 12.78678829, nan, nan, nan, nan, nan, nan], [ 9.80705187, 14.23670334, nan, nan, nan, nan, nan, nan], ... [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - median_volume_diameter(time, site)float647.077 0.0 nan nan ... nan nan nan
- long_name :
- Median volume diameter, assuming an ideal Marshall-Palmer type distribution
- units :
- mm
- _FillValue :
- -999999.0
- source :
- gucldM1.b1
array([[ 7.0774117 , 0. , nan, nan, nan, nan, nan, nan], [ 7.22024515, 0. , nan, nan, nan, nan, nan, nan], [16.90189689, 0. , nan, nan, nan, nan, nan, nan], [ 1.85330668, 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [13.5465818 , 2.40557567, nan, nan, nan, nan, nan, nan], [ 9.87035583, 0. , nan, nan, nan, nan, nan, nan], [ 9.66985622, 5.00972444, nan, nan, nan, nan, nan, nan], [ 9.32413797, 7.30939663, nan, nan, nan, nan, nan, nan], [ 9.66265531, 6.56595044, nan, nan, nan, nan, nan, nan], ... [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - liquid_water_distribution_mean(time, site)float647.714 0.0 nan nan ... nan nan nan
- long_name :
- Liquid water distribution mean, assuming an ideal Marshall-Palmer type distribution
- units :
- mm
- _FillValue :
- -999999.0
- source :
- gucldM1.b1
array([[ 7.7138003 , 0. , nan, nan, nan, nan, nan, nan], [ 7.86947741, 0. , nan, nan, nan, nan, nan, nan], [18.42168531, 0. , nan, nan, nan, nan, nan, nan], [ 2.0199528 , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [14.76466685, 2.62188092, nan, nan, nan, nan, nan, nan], [10.75788097, 0. , nan, nan, nan, nan, nan, nan], [10.53935268, 5.46018994, nan, nan, nan, nan, nan, nan], [10.16254684, 7.96664444, nan, nan, nan, nan, nan, nan], [10.53150527, 7.15635012, nan, nan, nan, nan, nan, nan], ... [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - number_density_drops(time, site, particle_size)float640.0 0.0 258.2 722.4 ... nan nan nan
- long_name :
- Number density of drops of the diameter corresponding to a particular drop size class per unit volume
- units :
- 1/(m^3 mm)
- _FillValue :
- -999999.0
- source :
- gucldM1.b1
array([[[ 0. , 0. , 258.18399745, ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ 0. , 0. , 181.54513377, ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ nan, nan, nan, ..., nan, nan, nan], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - diameter_min(time, site)float640.8463 0.0 nan nan ... nan nan nan
- long_name :
- Diameter of smallest drop observed
- units :
- mm
- _FillValue :
- -999999.0
- source :
- gucldM1.b1
array([[0.84628668, 0. , nan, nan, nan, nan, nan, nan], [1.21076004, 0. , nan, nan, nan, nan, nan, nan], [1.74916673, 0. , nan, nan, nan, nan, nan, nan], [0.18645334, 0. , nan, nan, nan, nan, nan, nan], [0. , 0. , nan, nan, nan, nan, nan, nan], [1.74166673, 0.59488001, nan, nan, nan, nan, nan, nan], [2.005 , 0. , nan, nan, nan, nan, nan, nan], [2.02999997, 1.09824002, nan, nan, nan, nan, nan, nan], [1.56000006, 1.56000006, nan, nan, nan, nan, nan, nan], [1.56000006, 1.56000006, nan, nan, nan, nan, nan, nan], ... [0. , 0. , nan, nan, nan, nan, nan, nan], [0. , 0. , nan, nan, nan, nan, nan, nan], [0. , 0. , nan, nan, nan, nan, nan, nan], [0. , 0. , nan, nan, nan, nan, nan, nan], [0. , 0. , nan, nan, nan, nan, nan, nan], [0. , 0. , nan, nan, nan, nan, nan, nan], [0. , 0. , nan, nan, nan, nan, nan, nan], [0. , 0. , nan, nan, nan, nan, nan, nan], [0. , 0. , nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - diameter_max(time, site)float6412.67 0.0 nan nan ... nan nan nan
- long_name :
- Diameter of largest drop observed
- units :
- mm
- _FillValue :
- -999999.0
- source :
- gucldM1.b1
array([[12.67166667, 0. , nan, nan, nan, nan, nan, nan], [15.56333333, 0. , nan, nan, nan, nan, nan, nan], [32.80541667, 0. , nan, nan, nan, nan, nan, nan], [ 3.52 , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [28.76666667, 5.49166667, nan, nan, nan, nan, nan, nan], [23.98 , 0. , nan, nan, nan, nan, nan, nan], [23.77 , 12.63 , nan, nan, nan, nan, nan, nan], [22.93 , 19.89 , nan, nan, nan, nan, nan, nan], [24.87666667, 18.33666667, nan, nan, nan, nan, nan, nan], ... [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ 0. , 0. , nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan]]) - beam_number(time, site)float64-9.999e+03 ... -9.999e+03
- long_name :
- Beam number
- units :
- 1
- _FillValue :
- -999999.0
- source :
- guc915rwpprecipmomenthighM1.a0
array([[-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], ... [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.]]) - beam_tilt_angle(time, site)float64-9.999e+03 ... -9.999e+03
- long_name :
- Beam tilt angle from the vertical
- units :
- degree
- _FillValue :
- -999999.0
- source :
- guc915rwpprecipmomenthighM1.a0
array([[-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], ... [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.]]) - beam_azimuth_angle(time, site)float64-9.999e+03 ... -9.999e+03
- long_name :
- Beam azimuth angle from true north
- units :
- degree
- _FillValue :
- -999999.0
- source :
- guc915rwpprecipmomenthighM1.a0
array([[-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], ... [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.]]) - doppler_velocity(time, height, site)float64-9.999e+03 ... -9.999e+03
- long_name :
- Doppler velocity in beam direction
- units :
- m/s
- _FillValue :
- -999999.0
- source :
- guc915rwpprecipmomenthighM1.a0
array([[[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., ... ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]]]) - spectral_width(time, height, site)float64-9.999e+03 ... -9.999e+03
- long_name :
- Width of the spectral peak
- units :
- m/s
- _FillValue :
- -999999.0
- source :
- guc915rwpprecipmomenthighM1.a0
array([[[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., ... ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]]]) - spectral_shape_score(time, height, site)float64-9.999e+03 ... -9.999e+03
- long_name :
- Score between 0 (bad) and 1 (good) that evaluates the shape of the spectral peak
- units :
- 1
- _FillValue :
- -999999.0
- source :
- guc915rwpprecipmomenthighM1.a0
array([[[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., ... ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]]]) - signal_power(time, height, site)float64-9.999e+03 ... -9.999e+03
- long_name :
- Magnitude of the spectral peak
- units :
- 1
- _FillValue :
- -999999.0
- source :
- guc915rwpprecipmomenthighM1.a0
array([[[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., ... ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]]]) - signal_to_other_ratio(time, height, site)float64-9.999e+03 ... -9.999e+03
- long_name :
- Signal of the selected peak to the signal of the largest other peak
- units :
- dB*100
- _FillValue :
- -999999.0
- source :
- guc915rwpprecipmomenthighM1.a0
array([[[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., ... ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]]]) - significance(time, height, site)float64-9.999e+03 ... -9.999e+03
- long_name :
- Significance score between 0 (bad) and 1 (good)
- units :
- 1
- _FillValue :
- -999999.0
- source :
- guc915rwpprecipmomenthighM1.a0
array([[[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., ... ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]]]) - noise_power(time, height, site)float64-9.999e+03 ... -9.999e+03
- long_name :
- Power of the noise floor
- units :
- 1
- _FillValue :
- -999999.0
- source :
- guc915rwpprecipmomenthighM1.a0
array([[[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., ... ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]]]) - dc_power(time, height, site)float64-9.999e+03 ... -9.999e+03
- long_name :
- Spectral density at zero Doppler shift
- units :
- 1
- _FillValue :
- -999999.0
- source :
- guc915rwpprecipmomenthighM1.a0
array([[[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., ... ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]]]) - multi_peak_steering_velocity(time, height, site)float64-9.999e+03 ... -9.999e+03
- long_name :
- Multi-peak steering velocity
- units :
- m/s
- _FillValue :
- -999999.0
- source :
- guc915rwpprecipmomenthighM1.a0
array([[[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., ... ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]]]) - multi_peak_average_significance(time, height, site)float64-9.999e+03 ... -9.999e+03
- long_name :
- Multi-peak average significance
- units :
- 1
- _FillValue :
- -999999.0
- source :
- guc915rwpprecipmomenthighM1.a0
array([[[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., ... ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]]]) - multi_peak_cumulative_significance(time, height, site)float64-9.999e+03 ... -9.999e+03
- long_name :
- Multi-peak cumulative significance
- units :
- 1
- _FillValue :
- -999999.0
- source :
- guc915rwpprecipmomenthighM1.a0
array([[[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., ... ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]]]) - multi_peak_strongest_percentage(time, height, site)float64-9.999e+03 ... -9.999e+03
- long_name :
- Multi-peak strongest percentage
- units :
- %
- _FillValue :
- -999999.0
- source :
- guc915rwpprecipmomenthighM1.a0
array([[[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., ... ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]]]) - multi_peak_partial_bonus_begin(time, height, site)float64-9.999e+03 ... -9.999e+03
- long_name :
- Multi-peak partial bonus begin
- units :
- m/s
- _FillValue :
- -999999.0
- source :
- guc915rwpprecipmomenthighM1.a0
array([[[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., ... ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]]]) - multi_peak_partial_bonus_end(time, height, site)float64-9.999e+03 ... -9.999e+03
- long_name :
- Multi-peak partial bonus end
- units :
- m/s
- _FillValue :
- -999999.0
- source :
- guc915rwpprecipmomenthighM1.a0
array([[[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., ... ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]]]) - multi_peak_full_bonus_begin(time, height, site)float64-9.999e+03 ... -9.999e+03
- long_name :
- Multi-peak full bonus begin
- units :
- m/s
- _FillValue :
- -999999.0
- source :
- guc915rwpprecipmomenthighM1.a0
array([[[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., ... ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]]]) - multi_peak_full_bonus_end(time, height, site)float64-9.999e+03 ... -9.999e+03
- long_name :
- Multi-peak full bonus end
- units :
- m/s
- _FillValue :
- -999999.0
- source :
- guc915rwpprecipmomenthighM1.a0
array([[[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., ... ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]]]) - cluster_average_significance(time, height, site)float64-9.999e+03 ... -9.999e+03
- long_name :
- Cluster average significance
- units :
- 1
- _FillValue :
- -999999.0
- source :
- guc915rwpprecipmomenthighM1.a0
array([[[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., ... ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]]]) - cluster_cumulative_significance(time, height, site)float64-9.999e+03 ... -9.999e+03
- long_name :
- Cluster cumulative significance
- units :
- 1
- _FillValue :
- -999999.0
- source :
- guc915rwpprecipmomenthighM1.a0
array([[[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., ... ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]]]) - cluster_consistency_rank(time, height, site)float64-9.999e+03 ... -9.999e+03
- long_name :
- Cluster consistency rank
- units :
- 1
- _FillValue :
- -999999.0
- source :
- guc915rwpprecipmomenthighM1.a0
array([[[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., ... ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]]]) - rwp_quality_flags(time, height, site)float64-9.999e+03 ... -9.999e+03
- long_name :
- RWP Quality flag
- units :
- 1
- flag_masks :
- 1,2,4,8,16,32,64,128,256,512,1024
- flag_meanings :
- no_peak weak low_SNR too_strong bad_shape ground_clutter large_other too_narrow too_wide cluster_weak_A cluster_weak_C
- bit_1_description :
- No peak was found
- bit_2_description :
- Significance level of the signal is too low
- bit_3_description :
- Signal to noise ratio is too low
- bit_4_description :
- Signal to noise ratio is too high
- bit_5_description :
- Too low spectral shape score
- bit_6_description :
- Ground-clutter contamination
- bit_7_description :
- Signal to other ratio is too low
- bit_8_description :
- Spectral width is too narrow
- bit_9_description :
- Spectral width is too wide
- bit_10_description :
- Cluster average significance level is too low
- bit_11_description :
- Cluster cumulative significance level is too low
- source :
- guc915rwpprecipmomenthighM1.a0
array([[[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., ... ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]], [[-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], ..., [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.], [-9999., -9999., -9999., ..., -9999., -9999., -9999.]]]) - number_coherent_integration(time, site)float64-9.999e+03 ... -9.999e+03
- long_name :
- Number of coherent integrations combined into each I/Q sample pair
- units :
- count
- _FillValue :
- -999999.0
- source :
- guc915rwpprecipmomenthighM1.a0
array([[-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], ... [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.]]) - number_incoherent_integration(time, site)float64-9.999e+03 ... -9.999e+03
- long_name :
- Number of incoherent integrations combined into each I/Q sample pair
- units :
- count
- _FillValue :
- -999999.0
- source :
- guc915rwpprecipmomenthighM1.a0
array([[-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], ... [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.]]) - number_FFT_points(time, site)float64-9.999e+03 ... -9.999e+03
- long_name :
- Number of spectral points used for the Fast Fourier Transform
- units :
- count
- _FillValue :
- -999999.0
- source :
- guc915rwpprecipmomenthighM1.a0
array([[-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], ... [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.]]) - interpulse_period(time, site)float64-9.999e+03 ... -9.999e+03
- long_name :
- Period between two pulses
- units :
- ns
- _FillValue :
- -999999.0
- source :
- guc915rwpprecipmomenthighM1.a0
array([[-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], ... [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.]]) - clock_phase_position(time, site)float64-9.999e+03 ... -9.999e+03
- long_name :
- SIRP clock phase position
- units :
- 1
- _FillValue :
- -999999.0
- source :
- guc915rwpprecipmomenthighM1.a0
array([[-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], ... [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.]]) - clock_phase_good_count(time, site)float64-9.999e+03 ... -9.999e+03
- long_name :
- SIRP clock phase good-count
- units :
- 1
- _FillValue :
- -999999.0
- source :
- guc915rwpprecipmomenthighM1.a0
array([[-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], ... [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.], [-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.]]) - pres(time, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Pressure
- units :
- hPa
- _FillValue :
- -999999.0
- valid_min :
- 0.0
- valid_max :
- 1100.0
- valid_delta :
- 10.0
- resolution :
- 0.1
- standard_name :
- air_pressure
- source :
- gucsondewnpnM1.b1
array([[ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, ... nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [177712.79007975, nan, nan, nan, nan, nan, nan, nan], [136821.18875 , nan, nan, nan, nan, nan, nan, nan], [ 62991.653125 , nan, nan, nan, nan, nan, nan, nan]]) - tdry(time, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Dry Bulb Temperature
- units :
- degC
- _FillValue :
- -999999.0
- valid_min :
- -90.0
- valid_max :
- 50.0
- valid_delta :
- 10.0
- resolution :
- 0.1
- standard_name :
- air_temperature
- source :
- gucsondewnpnM1.b1
array([[ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, ... nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ -1864.09348551, nan, nan, nan, nan, nan, nan, nan], [ -6389.42951172, nan, nan, nan, nan, nan, nan, nan], [-17317.1153125 , nan, nan, nan, nan, nan, nan, nan]]) - dp(time, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Dewpoint Temperature
- units :
- degC
- _FillValue :
- -999999.0
- valid_min :
- -110.0
- valid_max :
- 50.0
- resolution :
- 0.1
- standard_name :
- dew_point_temperature
- source :
- gucsondewnpnM1.b1
array([[ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, ... nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ -5870.55179214, nan, nan, nan, nan, nan, nan, nan], [-12338.72182292, nan, nan, nan, nan, nan, nan, nan], [-22307.1984375 , nan, nan, nan, nan, nan, nan, nan]]) - wspd(time, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Wind Speed
- units :
- m/s
- _FillValue :
- -999999.0
- valid_min :
- 0.0
- valid_max :
- 100.0
- resolution :
- 0.1
- standard_name :
- wind_speed
- source :
- gucsondewnpnM1.b1
array([[ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], ... [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [1146.89177859, nan, nan, nan, nan, nan, nan, nan], [5390.36779948, nan, nan, nan, nan, nan, nan, nan], [9651.8240625 , nan, nan, nan, nan, nan, nan, nan]]) - deg(time, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Wind Direction
- units :
- degree
- _FillValue :
- -999999.0
- valid_min :
- 0.0
- valid_max :
- 360.0
- resolution :
- 1.0
- standard_name :
- wind_from_direction
- source :
- gucsondewnpnM1.b1
array([[ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, ... nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ 78823.91666667, nan, nan, nan, nan, nan, nan, nan], [103595.54666667, nan, nan, nan, nan, nan, nan, nan], [ 99429.48 , nan, nan, nan, nan, nan, nan, nan]]) - rh(time, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Relative Humidity
- units :
- %
- _FillValue :
- -999999.0
- valid_min :
- 0.0
- valid_max :
- 100.0
- resolution :
- 1.0
- standard_name :
- relative_humidity
- source :
- gucsondewnpnM1.b1
array([[ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], ... [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [10065.68155924, nan, nan, nan, nan, nan, nan, nan], [ 4648.71153646, nan, nan, nan, nan, nan, nan, nan], [ 6848.76585938, nan, nan, nan, nan, nan, nan, nan]]) - u_wind(time, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Eastward Wind Component
- units :
- m/s
- _FillValue :
- -999999.0
- valid_min :
- -75.0
- valid_max :
- 75.0
- valid_delta :
- 5.0
- resolution :
- 0.1
- calculation :
- (-1.0 * sin(wind direction) * wind speed)
- standard_name :
- eastward_wind
- source :
- gucsondewnpnM1.b1
array([[ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], ... [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ 458.04518716, nan, nan, nan, nan, nan, nan, nan], [1370.23122721, nan, nan, nan, nan, nan, nan, nan], [4552.12089844, nan, nan, nan, nan, nan, nan, nan]]) - v_wind(time, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Northward Wind Component
- units :
- m/s
- _FillValue :
- -999999.0
- valid_min :
- -75.0
- valid_max :
- 75.0
- valid_delta :
- 5.0
- resolution :
- 0.1
- calculation :
- (-1.0 * cos(wind direction) * wind speed)
- standard_name :
- northward_wind
- source :
- gucsondewnpnM1.b1
array([[ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], ... [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ -959.15482417, nan, nan, nan, nan, nan, nan, nan], [-5210.86289062, nan, nan, nan, nan, nan, nan, nan], [-8338.76898437, nan, nan, nan, nan, nan, nan, nan]]) - wstat(time, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Wind Status
- units :
- 1
- _FillValue :
- -999999.0
- source :
- gucsondewnpnM1.b1
array([[nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], ... [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [nan, nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan], [ 0., nan, nan, nan, nan, nan, nan, nan]]) - asc(time, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Ascent Rate
- units :
- m/s
- _FillValue :
- -999999.0
- valid_min :
- -10.0
- valid_max :
- 20.0
- valid_delta :
- 5.0
- resolution :
- 0.1
- source :
- gucsondewnpnM1.b1
array([[ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], ... [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [1512.22495524, nan, nan, nan, nan, nan, nan, nan], [1239.31066406, nan, nan, nan, nan, nan, nan, nan], [1230.47602539, nan, nan, nan, nan, nan, nan, nan]]) - lat(site)float64-9.999e+03 ... -9.999e+03
- long_name :
- North latitude
- units :
- degree_N
- _FillValue :
- -999999.0
- valid_min :
- -90
- valid_max :
- 90
- standard_name :
- latitude
- comment :
- Latitude of SAIL In-Situ Ground Station
array([-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.])
- lon(site)float64-9.999e+03 ... -9.999e+03
- long_name :
- East longitude
- units :
- degree_E
- _FillValue :
- -999999.0
- valid_min :
- -180
- valid_max :
- 180
- standard_name :
- longitude
- comment :
- Longitude of SAIL In-Situ Ground Station
array([-9999., -9999., -9999., -9999., -9999., -9999., -9999., -9999.])
- alt(time, site)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Altitude above mean sea level
- units :
- m
- _FillValue :
- -999999.0
- standard_name :
- altitude
- source :
- gucsondewnpnM1.b1
array([[ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, ... nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [ nan, nan, nan, nan, nan, nan, nan, nan], [1028387.49365234, nan, nan, nan, nan, nan, nan, nan], [1928635.96 , nan, nan, nan, nan, nan, nan, nan], [3502378.89 , nan, nan, nan, nan, nan, nan, nan]])
- command_line :
- Conventions :
- ARM-1.3 CF/Radial instrument_parameters
- process_version :
- dod_version :
- input_datastreams :
- site_id :
- platform_id :
- facility_id :
- data_level :
- location_description :
- datastream :
- references :
- See XPRECIPRADAR Instrument Handbook
- doi :
- 10.5439/1884520
- insitution :
- U.S. Department of Energy Atmospheric Radiation Measurement (ARM) Climate Research Facility
- comment :
- This is highly experimental and initial data. There are many known and unknown issues. Please do not use before contacting the Translator responsible scollis@anl.gov
- attributions :
- This data is collected by the ARM Climate Research facility. Radar system is operated by the radar engineering team radar@arm.gov and the data is processed by the precipitation radar products team. LP code courtesy of Scott Giangrande BNL.
- vap_name :
- RadCLss
- known_issues :
- False phidp jumps in insect regions. Still uses old Giangrande code. Issues with some snow below melting layer.
- developers :
- Joseph O'Brien, ANL. Maxwell Grover, ANL. Robert Jackson, ANL. Zachary Sherman, ANL.
- translator :
- https://www.arm.gov/capabilities/instruments/xprecipradar
- mentors :
- https://www.arm.gov/connect-with-arm/organization/instrument-mentors/list#xprecipradar
- source :
- Colorado State University's X-Band Precipitation Radar Plan Position Indicator data processed with Corrected Moments in Antenna Coordinates 2.0 (xprecipradarcmacppi.c1) (DOI: 10.5439/1883164)
- field_names :
- DBZ, VEL, WIDTH, ZDR, PHIDP, RHOHV, NCP, DBZhv, cbb_flag, sounding_temperature, signal_to_noise_ratio, velocity_texture, gate_id, simulated_velocity, corrected_velocity, unfolded_differential_phase, corrected_differential_phase, filtered_corrected_differential_phase, corrected_specific_diff_phase, filtered_corrected_specific_diff_phase, corrected_differential_reflectivity, corrected_reflectivity, height_over_iso0, specific_attenuation, path_integrated_attenuation, specific_differential_attenuation, path_integrated_differential_attenuation, rain_rate_A, snow_rate_ws2012, snow_rate_ws88diw, snow_rate_m2009_1, snow_rate_m2009_2, latitude, longitude, intensity_rt, accum_rtnrt, accum_nrt, accum_total_nrt, bucket_rt, bucket_nrt, pluvio_status, maintenance_flag, reset_flag, intensity_rtnrt, atmos_pressure, temp_mean, temp_std, rh_mean, rh_std, vapor_pressure_mean, vapor_pressure_std, wspd_arith_mean, wspd_vec_mean, wdir_vec_mean, wdir_vec_std, pwd_err_code, pwd_mean_vis_1min, pwd_mean_vis_10min, pwd_pw_code_inst, pwd_pw_code_15min, pwd_pw_code_1hr, pwd_precip_rate_mean_1min, pwd_cumul_rain, pwd_cumul_snow, org_precip_rate_mean, tbrg_precip_total, tbrg_precip_total_corr, precip_rate, weather_code, number_detected_particles, mor_visibility, snow_depth_intensity, class_size_width, fall_velocity_calculated, raw_spectrum, liquid_water_content, equivalent_radar_reflectivity, intercept_parameter, slope_parameter, median_volume_diameter, liquid_water_distribution_mean, number_density_drops, diameter_min, diameter_max, beam_number, beam_tilt_angle, beam_azimuth_angle, doppler_velocity, spectral_width, spectral_shape_score, signal_power, signal_to_other_ratio, significance, noise_power, dc_power, multi_peak_steering_velocity, multi_peak_average_significance, multi_peak_cumulative_significance, multi_peak_strongest_percentage, multi_peak_partial_bonus_begin, multi_peak_partial_bonus_end, multi_peak_full_bonus_begin, multi_peak_full_bonus_end, cluster_average_significance, cluster_cumulative_significance, cluster_consistency_rank, rwp_quality_flags, number_coherent_integration, number_incoherent_integration, number_FFT_points, interpulse_period, clock_phase_position, clock_phase_good_count, pres, tdry, dp, wspd, deg, rh, u_wind, v_wind, wstat, asc, alt, height, time, site, particle_size, raw_fall_velocity
- history :
8) Save the DataSet#
# define a filename
out_ds.to_netcdf('xprecipradarradclss.c2.' + DATE.replace('-', '') + '.000000.nc')

